Note
Click here to download the full example code
07.e 3D Data with sns.displot
Figure-level interface for drawing distribution plots onto a FacetGrid.
This script demonstrates how to use the figure-level function
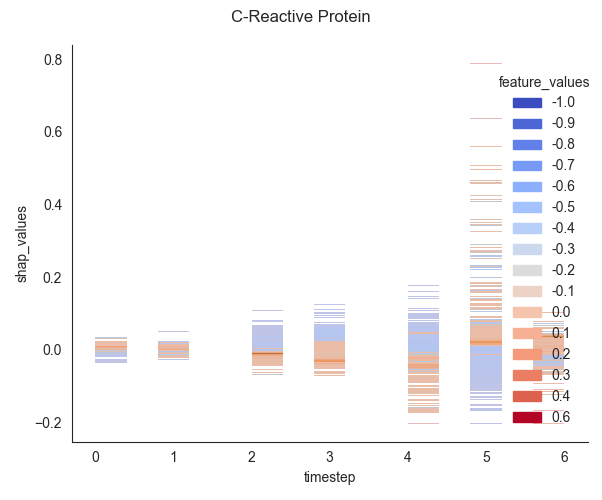
seaborn.displot to create a 2D histogram where the bins are
colored by the values of a third, continuous variable. 🎨
- The core technique involves:
Plotting two variables on the x and y axes to show their joint distribution (in this case, timestep and shap_values).
Using the hue parameter to map a third variable (feature_values) to the color of each bin, effectively visualizing a third dimension on a 2D plot.
Leveraging Seaborn’s hue_norm to correctly scale the colormap to the range of the third variable.
Out:
Unnamed: 0 sample timestep feature_values shap_values
count 7000.000000 7000.000000 7000.000000 7000.000000 7000.000000
mean 126003.000000 499.500000 3.000000 -0.570943 0.000091
std 72751.329885 288.695612 2.000143 0.410149 0.042353
min 21.000000 0.000000 0.000000 -1.000000 -0.204305
25% 63012.000000 249.750000 1.000000 -0.900000 -0.011614
50% 126003.000000 499.500000 3.000000 -0.800000 0.000515
75% 188994.000000 749.250000 5.000000 0.000000 0.012057
max 251985.000000 999.000000 6.000000 0.600000 0.789859
0. Computing... C-Reactive Protein
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main07_e_displot.py:155: UserWarning:
FigureCanvasAgg is non-interactive, and thus cannot be shown
22 import pandas as pd
23 import seaborn as sns
24 import matplotlib as mpl
25 import matplotlib.pyplot as plt
26
27 from pathlib import Path
28
29 sns.set_style(style="white")
30
31
32 def scalar_colormap(values, cmap, vmin, vmax):
33 """This method creates a colormap based on values.
34
35 Parameters
36 ----------
37 values : array-like
38 The values to create the corresponding colors
39
40 cmap : str
41 The colormap
42
43 vmin, vmax : float
44 The minimum and maximum possible values
45
46 Returns
47 -------
48 scalar colormap
49 """
50 # Create scalar mappable
51 norm = mpl.colors.Normalize(vmin=vmin, vmax=vmax, clip=True)
52 mapper = mpl.cm.ScalarMappable(norm=norm, cmap=cmap)
53 # Get color map
54 colormap = sns.color_palette([mapper.to_rgba(i) for i in values])
55 # Return
56 return colormap, norm
57
58 def scalar_palette(values, cmap, vmin, vmax):
59 """This method creates a colorpalette based on values.
60
61 Parameters
62 ----------
63 values : array-like
64 The values to create the corresponding colors
65
66 cmap : str
67 The colormap
68
69 vmin, vmax : float
70 The minimum and maximum possible values
71
72 Returns
73 -------
74 scalar colormap
75
76 """
77 # Create a matplotlib colormap from name
78 # cmap = sns.light_palette(cmap, reverse=False, as_cmap=True)
79 cmap = sns.color_palette(cmap, as_cmap=True)
80 # Normalize to the range of possible values from df["c"]
81 norm = mpl.colors.Normalize(vmin=vmin, vmax=vmax)
82 # Create a color dictionary (value in c : color from colormap)
83 colors = {}
84 for cval in values:
85 colors.update({cval: cmap(norm(cval))})
86 # Return
87 return colors, norm
88
89
90 # Load dataset
91 path = Path('../../datasets/shap')
92 data = pd.read_csv(path / 'shap.csv')
93 data = data[data.features.isin(['C-Reactive Protein'])]
94
95 # Since the colorbar is discrete, needs to round so that
96 # the amount of bins is small and therefore visible. Would
97 # it be possible to define a continuous colormap?
98 data.feature_values = data.feature_values.round(1)
99
100 # Show
101 print(data.describe())
102
103 # Configuration
104 cmap_name = 'coolwarm' # colormap name
105
106
107 # .. note:: The function displot calls the histplot function. However,
108 # the features allowed are count, frequency, probability or
109 # proportion, percent and density. Thus, the median cannot
110 # be computed.
111
112 # .. note:: The resulting colormap is discrete. Could it be continuous?
113
114 # Loop
115 for i, (name, df) in enumerate(data.groupby('features')):
116
117 # Info
118 print("%2d. Computing... %s" % (i, name))
119
120 # Get colormap
121 values = df.feature_values
122 cmap, norm = scalar_colormap(values=values,
123 cmap=cmap_name, vmin=values.min(),
124 vmax=values.max())
125
126 # Display displot
127 sns.displot(data=df, x='timestep', y='shap_values',
128 hue='feature_values', palette='coolwarm',
129 hue_norm=(values.min(), values.max()),
130 rug=False) # bins
131
132 """
133 # Display histplot
134 plt.figure()
135 sns.histplot(
136 data=df, x='timestep', y='shap_values',
137 discrete=(False, False),
138 hue='feature_values', palette=cmap_name,
139 hue_norm=(values.min(), values.max()),
140 cbar=False, cbar_kws=dict(shrink=.75),
141 #pthresh=.05, pmax=.9, bins=100
142 )
143 """
144
145 # Format figure
146 plt.suptitle(name)
147 plt.tight_layout()
148 plt.legend([], [], frameon=False)
149
150 # Show only first N
151 if int(i) > 2:
152 break
153
154 # Show
155 plt.show()
Total running time of the script: ( 0 minutes 2.860 seconds)