Note
Click here to download the full example code
07.d stats.2dbin with shap.csv
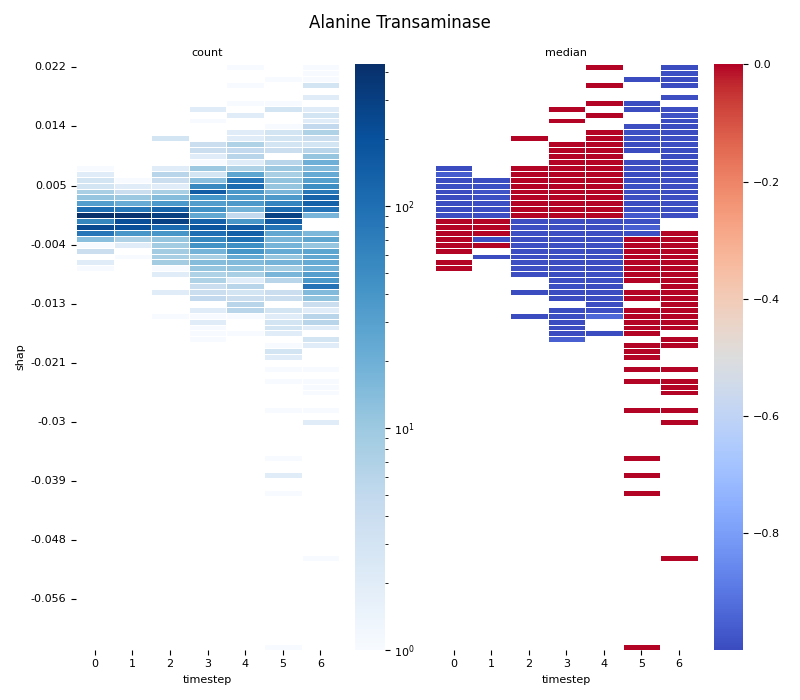
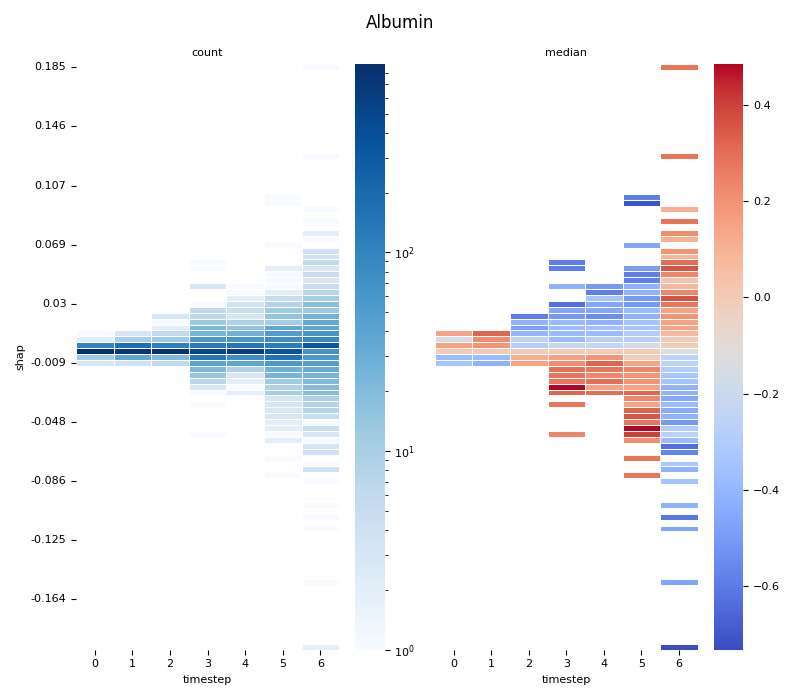
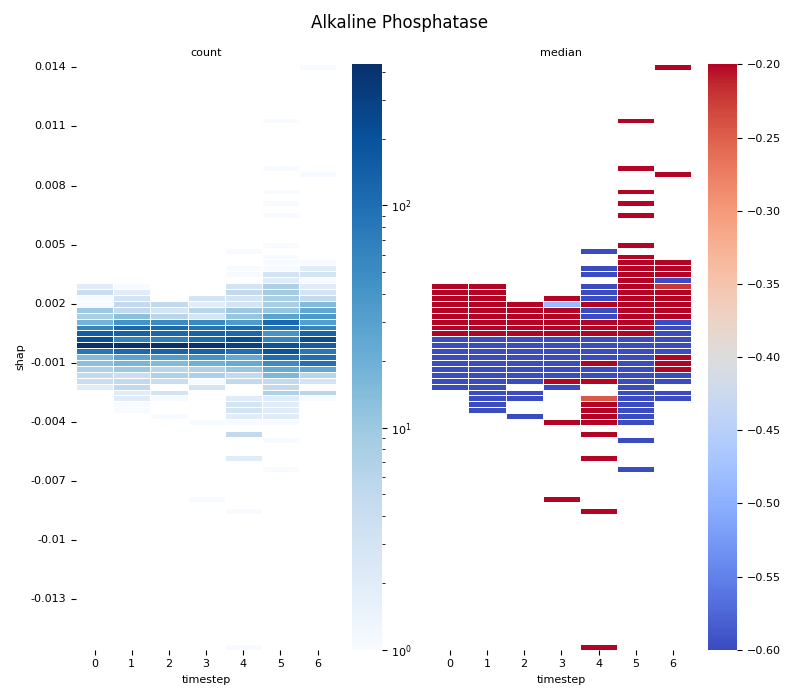
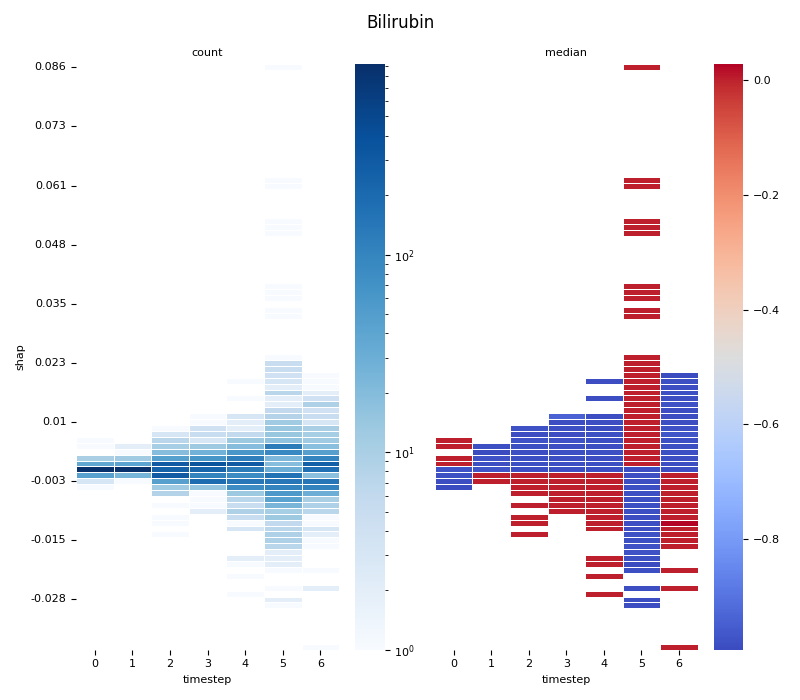
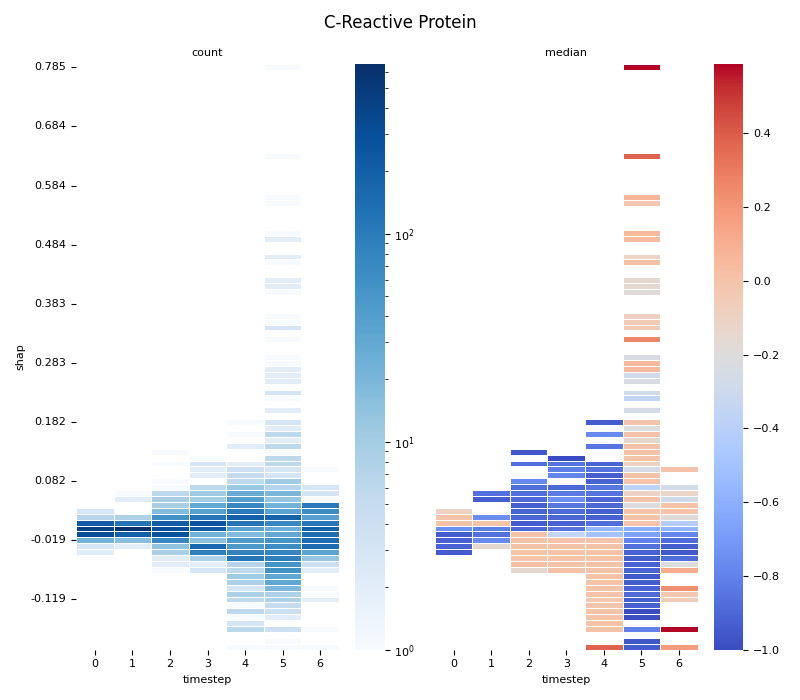
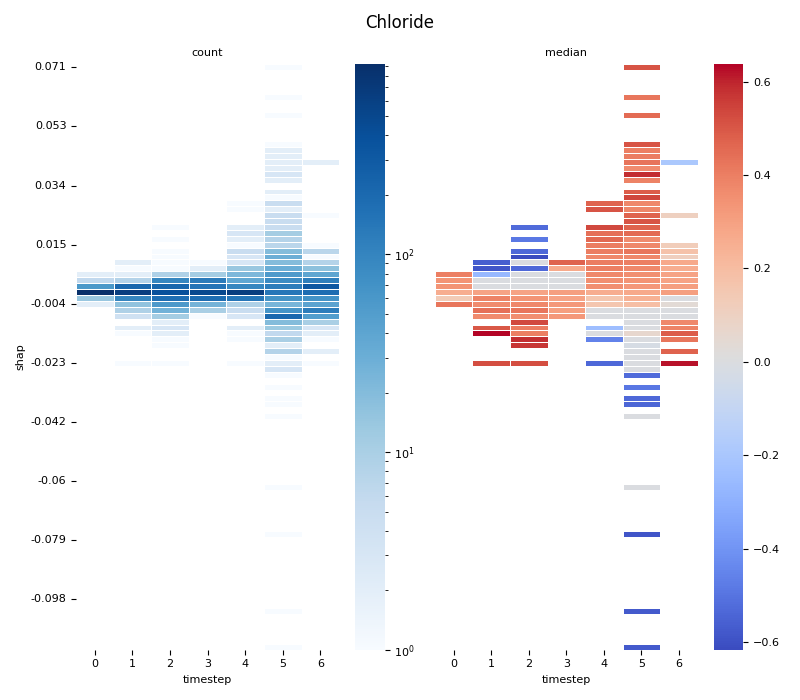
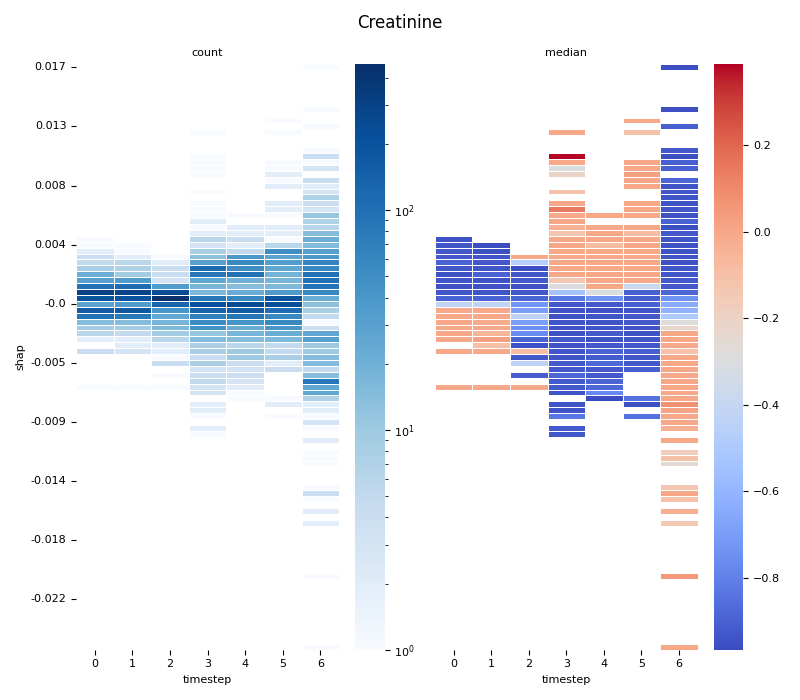
This script provides an advanced, per-feature analysis of time-series SHAP data, creating a dual-heatmap visualization to reveal complex interactions between feature values, their SHAP importance, and time. 📊
The workflow includes:
Per-Feature Processing: It iterates through individual features from a pre-computed SHAP dataset.
Statistical Binning: For each feature, it uses
scipy.stats.binned_statistic_2dto compute both thecountof data points and themedianof the original feature values for each cell in a 2D grid.Dual Heatmap Visualization: It plots two heatmaps side-by-side: one showing data density (with a log scale) and the other showing the median feature value (with a diverging colormap), allowing for direct comparison.
Out:
Unnamed: 0 sample timestep features feature_values shap_values
12 12 0 0 Creatinine 0.0 -0.001081
17 17 0 0 Chloride 0.0 -0.000858
21 21 0 0 C-Reactive Protein 0.0 0.010186
22 22 0 0 Albumin 0.0 0.000411
23 23 0 0 Alkaline Phosphatase 0.0 0.000486
27 27 0 0 Alanine Transaminase 0.0 -0.001809
28 28 0 0 Bilirubin 0.0 0.000500
48 48 0 1 Creatinine 0.0 -0.001033
53 53 0 1 Chloride 0.0 0.001109
57 57 0 1 C-Reactive Protein 0.0 0.005363
0. Computing... Alanine Transaminase
1. Computing... Albumin
2. Computing... Alkaline Phosphatase
3. Computing... Bilirubin
4. Computing... C-Reactive Protein
5. Computing... Chloride
6. Computing... Creatinine
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main07_d_2dbin_shap.py:173: UserWarning:
FigureCanvasAgg is non-interactive, and thus cannot be shown
23 # Libraries
24 import seaborn as sns
25 import pandas as pd
26 import numpy as np
27 import matplotlib as mpl
28 import matplotlib.pyplot as plt
29
30 from scipy import stats
31 from pathlib import Path
32 from matplotlib.colors import LogNorm
33
34 #plt.style.use('ggplot') # R ggplot style
35
36 # See https://matplotlib.org/devdocs/users/explain/customizing.html
37 mpl.rcParams['axes.titlesize'] = 8
38 mpl.rcParams['axes.labelsize'] = 8
39 mpl.rcParams['xtick.labelsize'] = 8
40 mpl.rcParams['ytick.labelsize'] = 8
41
42 # Constant
43 SNS_HEATMAP_CBAR_ARGS = {
44 'C-Reactive Protein': { 'vmin':-0.4, 'vmax':-0.2, 'center':-0.35 },
45 'Bilirubin': { 'vmin':-0.4, 'vmax':-0.2, 'center':-0.35 },
46 'Alanine Transaminase': {},
47 'Albumin': {},
48 'Alkaline Phosphatase': { 'vmin':-0.6, 'vmax':-0.2 },
49 'Bilirubin': {},
50 'C-Reactive Protein': {},
51 'Chloride': {},
52 }
53
54 # Load data
55 path = Path('../../datasets/shap/')
56 data = pd.read_csv(path / 'shap.csv')
57
58 # Filter
59 data = data[data.features.isin([
60 'Alanine Transaminase',
61 'Albumin',
62 'Alkaline Phosphatase',
63 'Bilirubin',
64 'C-Reactive Protein',
65 'Chloride',
66 'Creatinine'
67 ])]
68
69 # Show
70 print(data.head(10))
71
72 # figsize = (8,7) for 100 bins
73 # figsize = (8,3) for 50 bins
74 #
75 # .. note: The y-axis does not represent a continuous space,
76 # it is a discrete space where each tick is describing
77 # a bin.
78
79 # Loop
80 for i, (name, df) in enumerate(data.groupby('features')):
81
82 # Info
83 print("%2d. Computing... %s" % (i, name))
84
85 # Get variables
86 x = df.timestep
87 y = df.shap_values
88 z = df.feature_values
89 n = x.max()
90 vmin = z.min()
91 vmax = z.max()
92 nbins = 100
93 figsize = (8, 7)
94
95 # Create bins
96 binx = np.arange(x.min(), x.max()+2, 1) - 0.5
97 biny = np.linspace(y.min(), y.max(), nbins)
98
99 # Compute binned statistic (count)
100 r1 = stats.binned_statistic_2d(x=y, y=x, values=z,
101 statistic='count', bins=[biny, binx],
102 expand_binnumbers=False)
103
104 # Compute binned statistic (median)
105 r2 = stats.binned_statistic_2d(x=y, y=x, values=z,
106 statistic='median', bins=[biny, binx],
107 expand_binnumbers=False)
108
109 # Compute centres
110 x_center = (r1.x_edge[:-1] + r1.x_edge[1:]) / 2
111 y_center = (r1.y_edge[:-1] + r1.y_edge[1:]) / 2
112
113 # Flip
114 flip1 = np.flip(r1.statistic, 0)
115 flip2 = np.flip(r2.statistic, 0)
116
117 # Display
118 fig, axs = plt.subplots(nrows=1, ncols=2,
119 sharey=True, sharex=False, figsize=figsize)
120
121 sns.heatmap(flip1, annot=False, linewidth=0.5,
122 xticklabels=y_center.astype(int),
123 yticklabels=x_center.round(3)[::-1], # Because of flip
124 cmap='Blues', ax=axs[0], norm=LogNorm(),
125 cbar_kws={
126 #'label': 'value [unit]',
127 'use_gridspec': True,
128 'location': 'right'
129 }
130 )
131
132 sns.heatmap(flip2, annot=False, linewidth=0.5,
133 xticklabels=y_center.astype(int),
134 yticklabels=x_center.round(3)[::-1], # Because of flip
135 cmap='coolwarm', ax=axs[1], zorder=1,
136 **SNS_HEATMAP_CBAR_ARGS.get(name, {}),
137 #vmin=vmin, vmax=vmax, center=center, robust=False,
138 cbar_kws={
139 #'label': 'value [unit]',
140 'use_gridspec': True,
141 'location': 'right'
142 }
143 )
144
145 # Configure ax0
146 axs[0].set_title('count')
147 axs[0].set_xlabel('timestep')
148 axs[0].set_ylabel('shap')
149 axs[0].locator_params(axis='y', nbins=10)
150
151 # Configure ax1
152 axs[1].set_title('median')
153 axs[1].set_xlabel('timestep')
154 #axs[1].set_ylabel('shap')
155 axs[1].locator_params(axis='y', nbins=10)
156 axs[1].tick_params(axis=u'y', which=u'both', length=0)
157 # axs[1].invert_yaxis()
158
159 # Identify zero crossing
160 #zero_crossing = np.where(np.diff(np.sign(biny)))[0]
161 # Display line on that index (not exactly 0 though)
162 #plt.axhline(y=len(biny) - zero_crossing, color='lightgray', linestyle='--')
163
164 # Generic
165 plt.suptitle(name)
166 plt.tight_layout()
167
168 # Show only first N
169 if int(i) > 5:
170 break
171
172 # Show
173 plt.show()
Total running time of the script: ( 0 minutes 5.422 seconds)