Note
Click here to download the full example code
06.b sns.heatmap for CRI v2
Plot rectangular data as a color-encoded matrix.
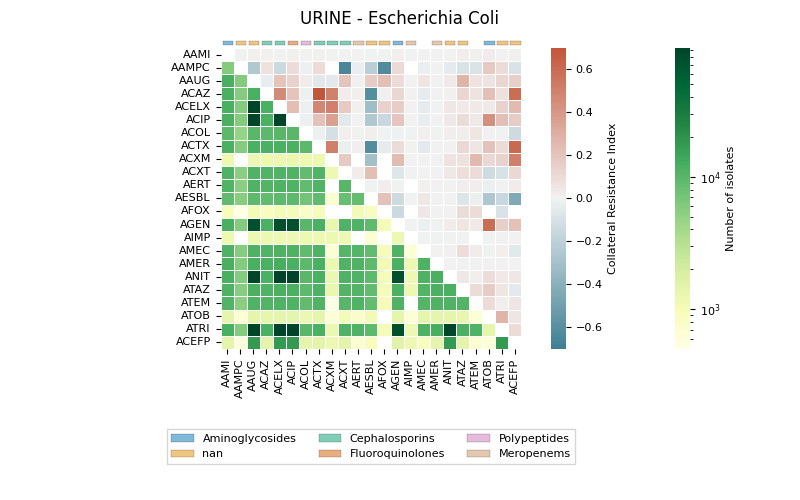
The generates a heatmap visualization for a dataset related to collateral sensitivity. It uses the Seaborn library to plot the rectangular data as a color-encoded matrix. The code loads the data from a CSV file, creates mappings for categories and colors, and then plots the heatmap using the loaded data and color maps. It also includes annotations, colorbar axes, category patches, legend elements, and formatting options to enhance the visualization.
16 # Libraries
17 import numpy as np
18 import pandas as pd
19 import seaborn as sns
20 import matplotlib as mpl
21 import matplotlib.pyplot as plt
22
23 from pathlib import Path
24 from matplotlib.patches import Patch
25 from matplotlib.colors import LogNorm
26 from matplotlib.patches import Rectangle
27
28 # See https://matplotlib.org/devdocs/users/explain/customizing.html
29 mpl.rcParams['axes.titlesize'] = 8
30 mpl.rcParams['axes.labelsize'] = 8
31 mpl.rcParams['xtick.labelsize'] = 8
32 mpl.rcParams['ytick.labelsize'] = 8
33
34 try:
35 __file__
36 TERMINAL = True
37 except:
38 TERMINAL = False
Lets load the data and create color mapping variables
43 # Load data
44 path = Path('../../datasets/collateral-sensitivity/sample')
45 data = pd.read_csv(path / 'matrix.csv', index_col=0)
46 abxs = pd.read_csv(path / 'categories.csv', index_col=0)
47
48 # Create dictionary to map category to color
49 labels = abxs.category
50 palette = sns.color_palette('colorblind', labels.nunique())
51 lookup = dict(zip(labels.unique(), palette))
52
53 # Create dictionary to map code to category
54 code2cat = dict(zip(abxs.antimicrobial_code, abxs.category))
55
56 # Create colors
57 colors = data.columns.to_series().map(code2cat).map(lookup)
Lets see the data
62 if TERMINAL:
63 print("\nData:")
64 print(data)
65 data.iloc[:7,:7]
Lets see the antimicrobials
69 if TERMINAL:
70 print("\nAntimicrobials:")
71 print(abxs)
72 abxs
Lets create some variables.
78 # Create color maps
79 cmapu = sns.color_palette("YlGn", as_cmap=True)
80 cmapl = sns.diverging_palette(220, 20, as_cmap=True)
81
82 # Create triangular matrices
83 masku = np.triu(np.ones_like(data))
84 maskl = np.tril(np.ones_like(data))
Let’s display a heatmap
91 # Draw (heatmap)
92 fig, axs = plt.subplots(nrows=1, ncols=1,
93 sharey=False, sharex=False, figsize=(8, 5)
94 )
95
96 # Display
97 r1 = sns.heatmap(data=data, cmap=cmapu, mask=masku, ax=axs,
98 annot=False, linewidth=0.5, norm=LogNorm(),
99 annot_kws={"size": 8}, square=True, vmin=0,
100 cbar_kws={'label': 'Number of isolates'})
101
102 r2 = sns.heatmap(data=data, cmap=cmapl, mask=maskl, ax=axs,
103 annot=False, linewidth=0.5, vmin=-0.7, vmax=0.7,
104 center=0, annot_kws={"size": 8}, square=True,
105 xticklabels=True, yticklabels=True,
106 cbar_kws={'label': 'Collateral Resistance Index'})
107
108 # Create patches for categories
109 category_patches = []
110 for i in axs.get_xticklabels():
111 try:
112 x, y = i.get_position()
113 c = colors.to_dict().get(i.get_text(), 'k')
114 #i.set_color(c) # for testing
115
116 # Add patch.
117 category_patches.append(
118 Rectangle((x-0.35, y-0.5), 0.8, 0.3, edgecolor='k',
119 facecolor=c, fill=True, lw=0.25, alpha=0.5, zorder=1000,
120 transform=axs.transData
121 )
122 )
123 except Exception as e:
124 print(i.get_text(), e)
125
126 # Add category rectangles
127 fig.patches.extend(category_patches)
128
129 # Create legend elements
130 legend_elements = [
131 Patch(facecolor=v, edgecolor='k',
132 fill=True, lw=0.25, alpha=0.5, label=k)
133 for k, v in lookup.items()
134 ]
135
136 # Add legend
137 axs.legend(handles=legend_elements, loc='upper center',
138 ncol=3, bbox_to_anchor=(0.5, -0.25), fontsize=8,
139 fancybox=False, shadow=False)
140
141 # Format
142 plt.suptitle('URINE - Escherichia Coli')
143 #plt.subplots_adjust(right=0.2)
144 plt.tight_layout()
145
146 # Show
147 plt.show()
Out:
AMEC Invalid RGBA argument: nan
ATEM Invalid RGBA argument: nan
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_b_heatmap.py:147: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
Total running time of the script: ( 0 minutes 0.533 seconds)