Note
Click here to download the full example code
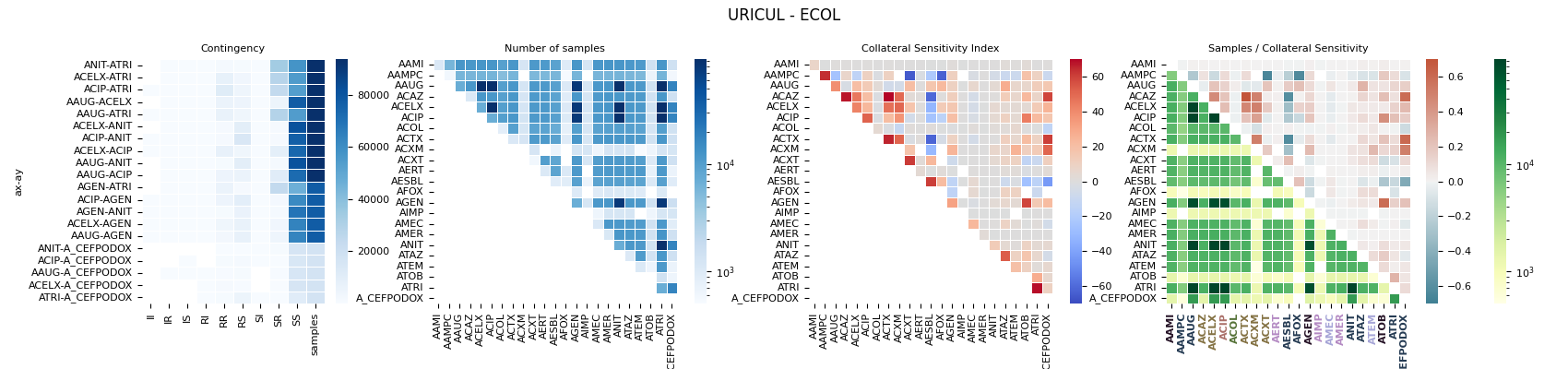
06.c Collateral Sensitivity Index (CRI)
This script creates a sophisticated, multi-panel visualization for pre-computed Collateral Sensitivity Index (CRI) data, designed for in-depth analysis of drug interactions.
The workflow includes:
Data Loading: It ingests pre-processed CRI and sample frequency data from CSV files.
Multi-Heatmap Layout: It generates several
seabornheatmaps: one for the CRI (using a diverging colormap), another for sample counts (with a log scale), and a composite heatmap combining both metrics in its upper and lower triangles.Categorical Annotation: It enhances the final plot by adding color-coded labels to the axes based on antibiotic categories, creating a dense, information-rich figure.
Note
Since the computation of the Collateral Sensitivity Index is quite computationally expensive, the results are saved into a .csv file so that can be easily loaded and displayed. This script shows a very basic graph of such information.
27 # Libraries
28 import numpy as np
29 import pandas as pd
30 import seaborn as sns
31 import matplotlib as mpl
32 import matplotlib.pyplot as plt
33
34 #
35 from pathlib import Path
36 from itertools import combinations
37 from matplotlib.colors import LogNorm, Normalize
38
39 # See https://matplotlib.org/devdocs/users/explain/customizing.html
40 mpl.rcParams['axes.titlesize'] = 8
41 mpl.rcParams['axes.labelsize'] = 8
42 mpl.rcParams['xtick.labelsize'] = 8
43 mpl.rcParams['ytick.labelsize'] = 8
44
45 try:
46 __file__
47 TERMINAL = True
48 except:
49 TERMINAL = False
50
51
52 # --------------------------------
53 # Methods
54 # --------------------------------
55 def _check_ax_ay_equal(ax, ay):
56 return ax==ay
57
58 def _check_ax_ay_greater(ax, ay):
59 return ax>ay
60
61 # --------------------------------
62 # Constants
63 # --------------------------------
64 # Possible cmaps
65 # https://r02b.github.io/seaborn_palettes/
66 # Diverging: coolwarm, RdBu_r, vlag
67 # Others: bone, gray, pink, twilight
68 cmap0 = 'coolwarm'
69 cmap1 = sns.light_palette("seagreen", as_cmap=True)
70 cmap2 = sns.color_palette("light:b", as_cmap=True)
71 cmap3 = sns.color_palette("vlag", as_cmap=True)
72 cmap4 = sns.diverging_palette(220, 20, as_cmap=True)
73 cmap5 = sns.color_palette("ch:s=.25,rot=-.25", as_cmap=True) # no
74 cmap6 = sns.color_palette("light:#5A9", as_cmap=True)
75 cmap7 = sns.light_palette("#9dedcc", as_cmap=True)
76 cmap8 = sns.color_palette("YlGn", as_cmap=True)
77
78 # Figure size
79 figsize = (17, 4)
Let’s load the data
85 # Load data
86 path = Path('../../datasets/collateral-sensitivity/20230525-135511')
87 data = pd.read_csv(path / 'contingency.csv')
88 abxs = pd.read_csv(path / 'categories.csv')
89
90 # Format data
91 data = data.set_index(['specimen', 'o', 'ax', 'ay'])
92 data.RR = data.RR.fillna(0).astype(int)
93 data.RS = data.RS.fillna(0).astype(int)
94 data.SR = data.SR.fillna(0).astype(int)
95 data.SS = data.SS.fillna(0).astype(int)
96 data['samples'] = data.RR + data.RS + data.SR + data.SS
97
98 #data['samples'] = data.iloc[:, :4].sum(axis=1)
99
100 def filter_top_pairs(df, n=5):
101 """Filter top n (Specimen, Organism) pairs."""
102 # Find top
103 top = df.groupby(level=[0, 1]) \
104 .samples.sum() \
105 .sort_values(ascending=False) \
106 .head(n)
107
108 # Filter
109 idx = pd.IndexSlice
110 a = top.index.get_level_values(0).unique()
111 b = top.index.get_level_values(1).unique()
112
113 # Return
114 return df.loc[idx[a, b, :, :]]
115
116 # Filter
117 data = filter_top_pairs(data, n=2)
118 data = data[data.samples > 500]
Lets see the data
122 if TERMINAL:
123 print("\n")
124 print("Number of samples: %s" % data.samples.sum())
125 print("Number of pairs: %s" % data.shape[0])
126 print("Data:")
127 print(data)
128 data.iloc[:7,:].dropna(axis=1, how='all')
Lets load the antimicrobial data and create color mapping variables
134 # Create dictionary to map category to color
135 labels = abxs.category
136 palette = sns.color_palette('Spectral', labels.nunique())
137 palette = sns.cubehelix_palette(labels.nunique(),
138 light=.9, dark=.1, reverse=True, start=1, rot=-2)
139 lookup = dict(zip(labels.unique(), palette))
140
141 # Create dictionary to map code to category
142 code2cat = dict(zip(abxs.antimicrobial_code, abxs.category))
Let’s display the information
148 # Loop
149 for i, df in data.groupby(level=[0, 1]):
150
151 # Drop level
152 df = df.droplevel(level=[0, 1])
153
154 # Check possible issues.
155 ax = df.index.get_level_values(0)
156 ay = df.index.get_level_values(1)
157 idx1 = _check_ax_ay_equal(ax, ay)
158 idx2 = _check_ax_ay_greater(ax, ay)
159
160 # Show
161 print("%25s. ax==ay => %5s | ax>ay => %5s" % \
162 (i, idx1.sum(), idx2.sum()))
163
164 # Re-index to have square matrix
165 abxs = set(ax) | set(ay)
166 index = pd.MultiIndex.from_product([abxs, abxs])
167
168 # Reformat MIS
169 mis = df['MIS'] \
170 .reindex(index, fill_value=np.nan) \
171 .unstack()
172
173 # Reformat samples
174 freq = df['samples'] \
175 .reindex(index, fill_value=0) \
176 .unstack()
177
178 # Combine in square matrix
179 m1 = mis.copy(deep=True).to_numpy()
180 m2 = freq.to_numpy()
181 il1 = np.tril_indices(mis.shape[1])
182 m1[il1] = m2.T[il1]
183 m = pd.DataFrame(m1,
184 index=mis.index, columns=mis.columns)
185
186 # .. note: This is the matrix that is used in previous
187 # samples to display the CRI and the count using
188 # the sns.heatmap function
189 # Save
190 #m.to_csv('%s'%str(i))
191
192 # Add frequency
193 top_n = df \
194 .sort_values('samples', ascending=False) \
195 .head(20).drop(columns='MIS') \
196 .dropna(axis=1, how='all')
197
198 # Draw
199 fig, axs = plt.subplots(nrows=1, ncols=4,
200 sharey=False, sharex=False, figsize=figsize,
201 gridspec_kw={'width_ratios': [2, 3, 3, 3.5]})
202
203 sns.heatmap(data=mis * 100, annot=False, linewidth=.5,
204 cmap='coolwarm', vmin=-70, vmax=70, center=0,
205 annot_kws={"size": 8}, square=True,
206 ax=axs[2], xticklabels=True, yticklabels=True)
207
208 sns.heatmap(data=freq, annot=False, linewidth=.5,
209 cmap='Blues', norm=LogNorm(),
210 annot_kws={"size": 8}, square=True,
211 ax=axs[1], xticklabels=True, yticklabels=True)
212
213 sns.heatmap(top_n,
214 annot=False, linewidth=0.5,
215 cmap='Blues', ax=axs[0], zorder=1,
216 vmin=None, vmax=None, center=None, robust=True,
217 square=False, xticklabels=True, yticklabels=True,
218 cbar_kws={
219 'use_gridspec': True,
220 'location': 'right'
221 }
222 )
223
224 # Display
225 masku = np.triu(np.ones_like(m))
226 maskl = np.tril(np.ones_like(m))
227 sns.heatmap(data=m, cmap=cmap8, mask=masku, ax=axs[3],
228 annot=False, linewidth=0.5, norm=LogNorm(),
229 annot_kws={"size": 8}, square=True, vmin=0)
230 sns.heatmap(data=m, cmap=cmap4, mask=maskl, ax=axs[3],
231 annot=False, linewidth=0.5, vmin=-0.7, vmax=0.7,
232 center=0, annot_kws={"size": 8}, square=True,
233 xticklabels=True, yticklabels=True)
234
235 # Configure axes
236 axs[0].set_title('Contingency')
237 axs[1].set_title('Number of samples')
238 axs[2].set_title('Collateral Sensitivity Index')
239 axs[3].set_title('Samples / Collateral Sensitivity')
240
241 # Add colors to xticklabels
242
243 #abxs = pd.read_csv('../../datasets/susceptibility-nhs/susceptibility-v0.0.1/antimicrobials.csv')##
244
245 #groups = dict(zip(abxs.antimicrobial_code, abxs.category))
246 #cmap = sns.color_palette("Spectral", abxs.category.nunique())
247 #colors = dict(zip(abxs.category, cmap))
248
249 # ------------------------------------------
250 # Add category colors on xtick labels
251 # ------------------------------------------
252 # Create colors
253 colors = m.columns.to_series().map(code2cat).map(lookup)
254
255 # Loop
256 for lbl in axs[3].get_xticklabels():
257 try:
258 x, y = lbl.get_position()
259 c = colors.to_dict().get(lbl.get_text(), 'k')
260 lbl.set_color(c)
261 lbl.set_weight('bold')
262
263 """
264 axs[3].annotate('', xy=(2000, 0),
265 #xytext=(0, -15 - axs[3].xaxis.labelpad),
266 xytext=(i.x, y)
267 xycoords=('data', 'axes fraction'),
268 textcoords='offset points',
269 ha='center', va='top',
270 bbox=dict(boxstyle='round', fc='none', ec='red'))
271 """
272 except Exception as e:
273 print(lbl.get_text(), e)
274
275 # Configure plot
276 plt.suptitle('%s - %s' % (i[0], i[1]))
277 plt.tight_layout()
278 plt.subplots_adjust(wspace=0.1)
279
280 # Exit loop
281 break
282
283 # Show
284 plt.show()
Out:
('URICUL', 'ECOL'). ax==ay => 18 | ax>ay => 0
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_c_collateral_sensitivity.py:284: UserWarning:
FigureCanvasAgg is non-interactive, and thus cannot be shown
Total running time of the script: ( 0 minutes 3.236 seconds)