Note
Click here to download the full example code
06.d Collateral Sensitivity Index (CRI)
Since the computation of the Collateral Sensitivity Index is quite computationally expensive, the results are saved into a .csv file so that can be easily loaded and displayed. This script shows a very basic graph of such information.
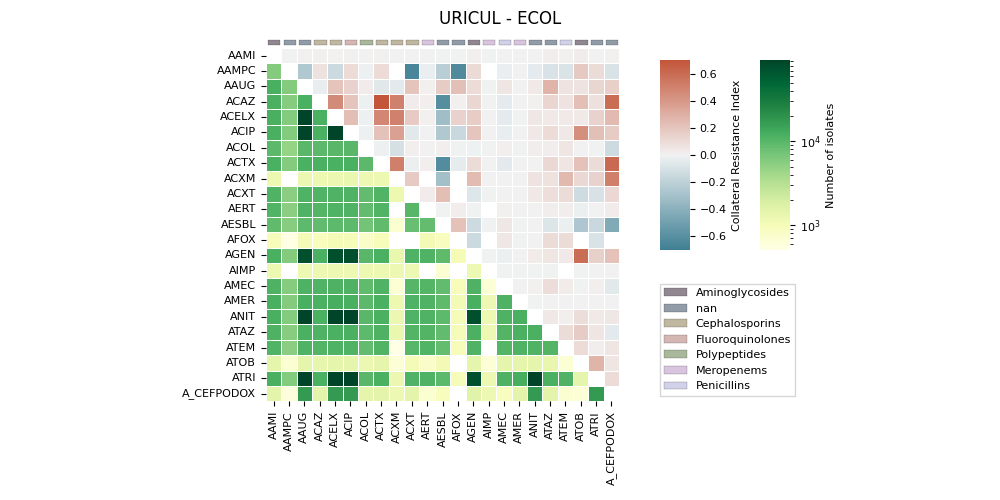
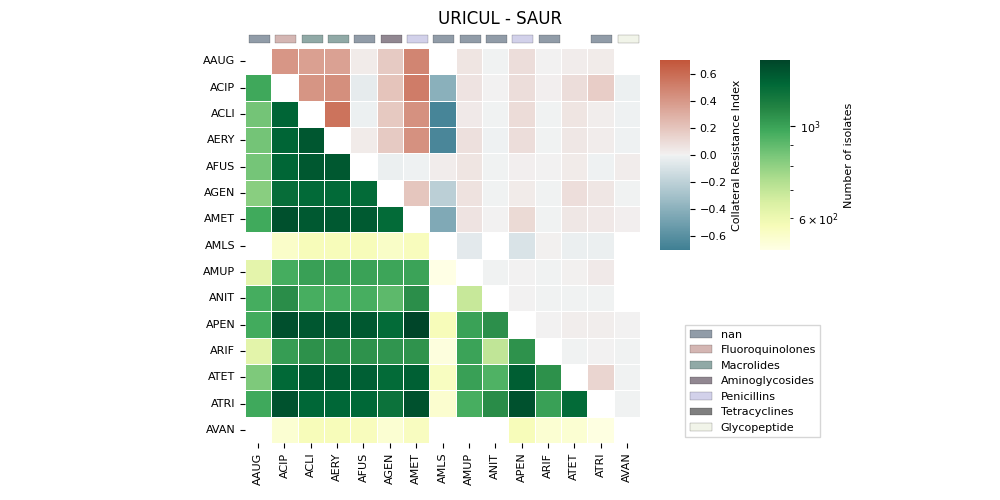
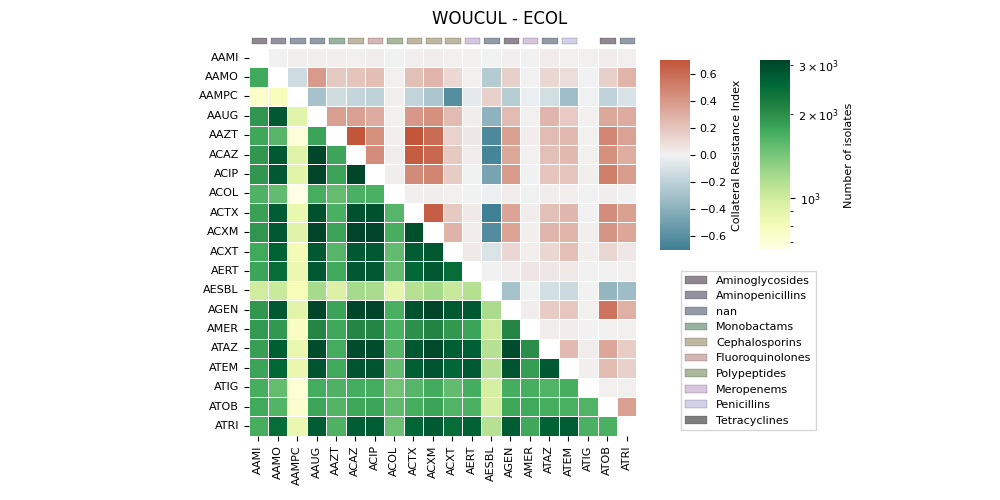
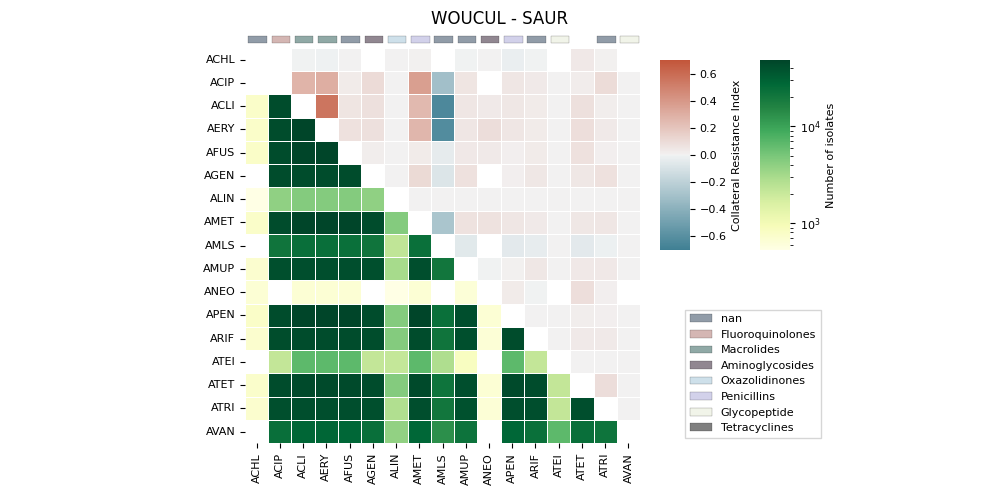
The generates a heatmap visualization for a dataset related to collateral sensitivity. It uses the Seaborn library to plot the rectangular data as a color-encoded matrix. The code loads the data from a CSV file, creates mappings for categories and colors, and then plots the heatmap using the loaded data and color maps. It also includes annotations, colorbar axes, category patches, legend elements, and formatting options to enhance the visualization.
19 # Libraries
20 import numpy as np
21 import pandas as pd
22 import seaborn as sns
23 import matplotlib as mpl
24 import matplotlib.pyplot as plt
25
26 # Specific libraries
27 from pathlib import Path
28 from matplotlib.patches import Patch
29 from matplotlib.patches import Rectangle
30 from matplotlib.colors import LogNorm, Normalize
31
32 # See https://matplotlib.org/devdocs/users/explain/customizing.html
33 mpl.rcParams['axes.titlesize'] = 8
34 mpl.rcParams['axes.labelsize'] = 8
35 mpl.rcParams['xtick.labelsize'] = 8
36 mpl.rcParams['ytick.labelsize'] = 8
37
38 try:
39 __file__
40 TERMINAL = True
41 except:
42 TERMINAL = False
43
44
45 # --------------------------------
46 # Methods
47 # --------------------------------
48 def _check_ax_ay_equal(ax, ay):
49 return ax==ay
50
51 def _check_ax_ay_greater(ax, ay):
52 return ax>ay
53
54 # --------------------------------
55 # Constants
56 # --------------------------------
57 # Figure size
58 figsize = (10, 5)
Let’s load the data
63 # Load data
64 path = Path('../../datasets/collateral-sensitivity/20230525-135511')
65 data = pd.read_csv(path / 'contingency.csv')
66 abxs = pd.read_csv(path / 'categories.csv')
67
68 # Format data
69 data = data.set_index(['specimen', 'o', 'ax', 'ay'])
70 data.RR = data.RR.fillna(0).astype(int)
71 data.RS = data.RS.fillna(0).astype(int)
72 data.SR = data.SR.fillna(0).astype(int)
73 data.SS = data.SS.fillna(0).astype(int)
74 data['samples'] = data.RR + data.RS + data.SR + data.SS
75
76 #data['samples'] = data.iloc[:, :4].sum(axis=1)
77
78 def filter_top_pairs(df, n=5):
79 """Filter top n (Specimen, Organism) pairs."""
80 # Find top
81 top = df.groupby(level=[0, 1]) \
82 .samples.sum() \
83 .sort_values(ascending=False) \
84 .head(n)
85
86 # Filter
87 idx = pd.IndexSlice
88 a = top.index.get_level_values(0).unique()
89 b = top.index.get_level_values(1).unique()
90
91 # Return
92 return df.loc[idx[a, b, :, :]]
93
94 # Filter
95 data = filter_top_pairs(data, n=2)
96 data = data[data.samples > 500]
Lets see the data
100 if TERMINAL:
101 print("\n")
102 print("Number of samples: %s" % data.samples.sum())
103 print("Number of pairs: %s" % data.shape[0])
104 print("Data:")
105 print(data)
106 data.iloc[:7,:].dropna(axis=1, how='all')
Lets load the antimicrobial data and create color mapping variables
111 # Create dictionary to map category to color
112 labels = abxs.category
113 palette = sns.color_palette('colorblind', labels.nunique())
114 palette = sns.cubehelix_palette(labels.nunique(),
115 light=.9, dark=.1, reverse=True, start=1, rot=-2)
116 lookup = dict(zip(labels.unique(), palette))
117
118 # Create dictionary to map code to category
119 code2cat = dict(zip(abxs.antimicrobial_code, abxs.category))
Let’s display the information
125 # Loop
126 for i, df in data.groupby(level=[0, 1]):
127
128 # Drop level
129 df = df.droplevel(level=[0, 1])
130
131 # Check possible issues.
132 ax = df.index.get_level_values(0)
133 ay = df.index.get_level_values(1)
134 idx1 = _check_ax_ay_equal(ax, ay)
135 idx2 = _check_ax_ay_greater(ax, ay)
136
137 # Show
138 print("%25s. ax==ay => %5s | ax>ay => %5s" % \
139 (i, idx1.sum(), idx2.sum()))
140
141 # Re-index to have square matrix
142 abxs = set(ax) | set(ay)
143 index = pd.MultiIndex.from_product([abxs, abxs])
144
145 # Reformat MIS
146 mis = df['MIS'] \
147 .reindex(index, fill_value=np.nan) \
148 .unstack()
149
150 # Reformat samples
151 freq = df['samples'] \
152 .reindex(index, fill_value=0) \
153 .unstack()
154
155 # Combine in square matrix
156 m1 = mis.copy(deep=True).to_numpy()
157 m2 = freq.to_numpy()
158 il1 = np.tril_indices(mis.shape[1])
159 m1[il1] = m2.T[il1]
160 m = pd.DataFrame(m1,
161 index=mis.index, columns=mis.columns)
162
163 # ------------------------------------------
164 # Display heatmaps
165 # ------------------------------------------
166 # Create color maps
167 cmapu = sns.color_palette("YlGn", as_cmap=True)
168 cmapl = sns.diverging_palette(220, 20, as_cmap=True)
169
170 # Masks
171 masku = np.triu(np.ones_like(m))
172 maskl = np.tril(np.ones_like(m))
173
174 # Draw figure
175 fig, axs = plt.subplots(nrows=1, ncols=1,
176 sharey=False, sharex=False, figsize=figsize)
177
178 # Create own colorbar axes
179 # Params are [left, bottom, width, height]
180 cbar_ax1 = fig.add_axes([0.66, 0.5, 0.03, 0.38])
181 cbar_ax2 = fig.add_axes([0.76, 0.5, 0.03, 0.38])
182
183 # Display
184 r1 = sns.heatmap(data=m, cmap=cmapu, mask=masku, ax=axs,
185 annot=False, linewidth=0.5, norm=LogNorm(),
186 annot_kws={"size": 8}, square=True, vmin=0,
187 cbar_ax=cbar_ax2,
188 cbar_kws={'label': 'Number of isolates'})
189
190 r2 = sns.heatmap(data=m, cmap=cmapl, mask=maskl, ax=axs,
191 annot=False, linewidth=0.5, vmin=-0.7, vmax=0.7,
192 center=0, annot_kws={"size": 8}, square=True,
193 xticklabels=True, yticklabels=True,
194 cbar_ax=cbar_ax1,
195 cbar_kws={'label': 'Collateral Resistance Index'})
196
197
198 # ------------------------------------------
199 # Add category rectangular patches
200 # ------------------------------------------
201 # Create colors
202 colors = m.columns.to_series().map(code2cat).map(lookup)
203
204 # Create patches for categories
205 category_patches = []
206 for lbl in axs.get_xticklabels():
207 try:
208 x, y = lbl.get_position()
209 c = colors.to_dict().get(lbl.get_text(), 'k')
210 # i.set_color(c) # for testing
211
212 # Add patch.
213 category_patches.append(
214 Rectangle((x - 0.35, y - 0.5), 0.8, 0.3, edgecolor='k',
215 facecolor=c, fill=True, lw=0.25, alpha=0.5,
216 zorder=1000, transform=axs.transData)
217 )
218 except Exception as e:
219 print(lbl.get_text(), e)
220
221 # Add category rectangles
222 fig.patches.extend(category_patches)
223
224
225 # ------------------------------------------
226 # Add category legend
227 # ------------------------------------------
228 # Unique categories
229 unique_categories = m.columns \
230 .to_series().map(code2cat).unique()
231
232 # Create legend elements
233 legend_elements = [
234 Patch(facecolor=lookup.get(k, 'k'), edgecolor='k',
235 fill=True, lw=0.25, alpha=0.5, label=k)
236 for k in unique_categories
237 ]
238
239 # Add legend
240 axs.legend(handles=legend_elements, loc='lower left',
241 ncol=1, bbox_to_anchor=(1.1, 0.00), fontsize=8,
242 fancybox=False, shadow=False)
243
244 # Configure plot
245 plt.suptitle('%s - %s' % (i[0], i[1]))
246 plt.tight_layout()
247 plt.subplots_adjust(left=-0.1, wspace=0.1)
248
249
250 # Show
251 plt.show()
Out:
('URICUL', 'ECOL'). ax==ay => 18 | ax>ay => 0
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_d_collateral_sensitivity.py:246: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
('URICUL', 'SAUR'). ax==ay => 0 | ax>ay => 0
ATET Invalid RGBA argument: nan
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_d_collateral_sensitivity.py:246: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
('WOUCUL', 'ECOL'). ax==ay => 0 | ax>ay => 0
ATIG Invalid RGBA argument: nan
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_d_collateral_sensitivity.py:246: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
('WOUCUL', 'SAUR'). ax==ay => 13 | ax>ay => 0
ATET Invalid RGBA argument: nan
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_d_collateral_sensitivity.py:246: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\matplotlib\plot_main06_d_collateral_sensitivity.py:251: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
Total running time of the script: ( 0 minutes 2.266 seconds)