Note
Go to the end to download the full example code
DRI - Example using MIMIC
In the field of healthcare, analyzing drug resistance in antimicrobial
use is crucial for understanding and combating the growing problem of
antibiotic resistance. One example is the Drug Resistance Index o DRI.
In this example, we compute such index using MIMIC, comprehensive,
a widely-used and freely available healthcare database that contains
de-identified electronic health records of over 60,000 intensive care unit
patients. Within MIMIC, researchers have access to rich information,
including patient demographics, clinical notes, laboratory results, and
medication records. This dataset provides the necessary data, susceptibility
and prescription information, to compute the drug resistance index.
Note
In MIMIC, the deidentification process for structured data
required the removal of dates. In particular, dates were shifted
into the future by a random offset for each individual patient in
a consistent manner to preserve intervals, resulting in stays which
occur sometime between the years 2100 and 2200. Time of day, day of
the week, and approximate seasonality were conserved during date
shifting.
26 # Define sphinx gallery configuration
27 # sphinx_gallery_thumbnail_number = 2
28
29 # Libraries
30 import sys
31 import warnings
32 import numpy as np
33 import pandas as pd
34 import seaborn as sns
35 import matplotlib as mpl
36
37 from pathlib import Path
38
39 try:
40 __file__
41 TERMINAL = True
42 except:
43 TERMINAL = False
44
45 # -------------------------
46 # Configuration
47 # -------------------------
48 # Params
49 rc = {
50 'axes.linewidth': 0.5,
51 'axes.labelsize': 9,
52 'axes.titlesize': 11,
53 'xtick.labelsize': 7,
54 'ytick.labelsize': 7,
55 }
56
57 # Configure seaborn style (context=talk)
58 sns.set_theme(style="white", rc=rc)
59
60 # Configure warnings
61 warnings.filterwarnings("ignore",
62 category=pd.errors.DtypeWarning)
63
64 # -------------------------------------------------------
65 # Constants
66 # -------------------------------------------------------
67 # Rename columns for susceptibility
68 rename_susceptibility = {
69 'chartdate': 'DATE',
70 'micro_specimen_id': 'LAB_NUMBER',
71 'spec_type_desc': 'SPECIMEN',
72 'org_name': 'MICROORGANISM',
73 'ab_name': 'ANTIMICROBIAL',
74 'interpretation': 'SENSITIVITY'
75 }
76
77 # Rename columns for prescriptions
78 rename_prescriptions = {
79 'drug': 'DRUG'
80 }
First, we need to load the susceptibility test data
87 # -----------------------------
88 # Load susceptibility test data
89 # -----------------------------
90 # Helper
91 subset = rename_susceptibility.values()
92
93 # Load data
94 path = Path('../../pyamr/datasets/mimic')
95 data1 = pd.read_csv(path / 'susceptibility.csv')
96
97 # Rename columns
98 data1 = data1.rename(columns=rename_susceptibility)
99
100 # Format data
101 data1 = data1[subset]
102 data1 = data1.dropna(subset=subset, how='any')
103 data1.DATE = pd.to_datetime(data1.DATE)
104 data1.SENSITIVITY = data1.SENSITIVITY.replace({
105 'S': 'sensitive',
106 'R': 'resistant',
107 'I': 'intermediate',
108 'P': 'pass'
109 })
112 data1.head(5)
Lets also load the prescriptions (could be also overall usage) data
118 # ----------------------------
119 # Load prescriptions
120 # ----------------------------
121 # Load prescription data (limited to first nrows).
122 data2 = pd.read_csv(path / 'prescriptions.csv', nrows=100000)
123 data2 = data2.rename(columns=rename_prescriptions)
124
125 # Format data
126 data2.DRUG = data2.DRUG.str.upper()
127 data2['DATE'] = pd.to_datetime(data2.starttime)
128 data2 = data2.dropna(subset=['DATE'], how='any')
129
130 # .. note:: We are only keeping those DRUGS which have
131 # the exact name of the antimicrobial tested
132 # in the susceptibility test record. There are
133 # also brand names that could/should be
134 # included
135
136 # Filter
137 data2 = data2[data2.DRUG.isin(data1.ANTIMICROBIAL.unique())]
140 print(data2)
subject_id hadm_id pharmacy_id starttime stoptime drug_type DRUG gsn ... form_rx dose_val_rx dose_unit_rx form_val_disp form_unit_disp doses_per_24_hrs route DATE
26 17868682 25218370 81568564 2167-05-24 20:00:00 2167-05-26 02:00:00 MAIN CEFAZOLIN 068632 ... NaN 2 g 1 BAG 3.0 IV 2167-05-24 20:00:00
48 17868682 24052239 59994266 2167-06-20 23:00:00 2167-06-22 11:00:00 MAIN VANCOMYCIN 043952 ... NaN 1000 mg 200 mL 2.0 IV 2167-06-20 23:00:00
58 17868682 24052239 39104292 2167-06-22 20:00:00 2167-06-25 12:00:00 MAIN VANCOMYCIN 009331 ... NaN 1500 mg 3 VIAL 2.0 IV 2167-06-22 20:00:00
60 17868682 24052239 1791946 2167-06-24 20:00:00 2167-06-25 12:00:00 MAIN VANCOMYCIN 009331 ... NaN 1500 mg 3 VIAL 2.0 IV 2167-06-24 20:00:00
76 12315540 24554730 25748315 2172-08-15 11:00:00 2172-08-16 20:00:00 MAIN LEVOFLOXACIN 029928 ... NaN 750 mg 1.5 TAB 0.0 PO 2172-08-15 11:00:00
... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ...
99154 15276693 21600683 24768410 2141-08-21 08:00:00 2141-08-26 18:00:00 MAIN LEVOFLOXACIN 46771.0 ... NaN 750 mg 1 TAB 0.0 PO/NG 2141-08-21 08:00:00
99218 10819462 26263456 61192290 2181-12-01 16:00:00 2181-12-02 15:00:00 MAIN CLINDAMYCIN 9344.0 ... NaN 600 mg 4 mL 3.0 IV 2181-12-01 16:00:00
99435 17442082 26038207 88544744 2135-08-07 15:00:00 2135-08-08 14:00:00 MAIN CEFEPIME 24095.0 ... NaN 2 g 1 VIAL 1.0 IV 2135-08-07 15:00:00
99511 17442082 20010297 74705027 2135-09-10 07:00:00 2135-09-10 11:00:00 MAIN CEFTRIAXONE 9162.0 ... NaN 1 gm 1 VIAL 1.0 IV 2135-09-10 07:00:00
99760 19103067 28239677 20279428 2124-06-02 04:00:00 2124-06-02 22:00:00 MAIN CLINDAMYCIN 9339.0 ... NaN 300 mg 2 CAP 3.0 PO/NG 2124-06-02 04:00:00
[2655 rows x 18 columns]
Lets rename the variables.
145 # Rename variables
146 susceptibility, prescriptions = data1, data2
147
148 # Show
149 if TERMINAL:
150 print("\nSusceptibility:")
151 print(susceptibility.head(10))
152 print("\nPrescriptions:")
153 print(prescriptions.head(10))
Now we need to create a summary table including the resistance
value, which will be computed using SARI and the usage which
will be computed manually. This summary table is required as a
preliminary step to compute the DRI.
162 # ------------------------
163 # Compute summary table
164 # ------------------------
165 # Libraries
166 from pyamr.core.sari import SARI
167
168 # Create sari instance
169 sari = SARI(groupby=[
170 susceptibility.DATE.dt.year,
171 #'SPECIMEN',
172 'MICROORGANISM',
173 'ANTIMICROBIAL',
174 'SENSITIVITY']
175 )
176
177 # Compute susceptibility summary table
178 smmry1 = sari.compute(susceptibility,
179 return_frequencies=True)
180
181 # .. note:: We are counting the number of rows as an indicator
182 # of prescriptions. However, it would be possible to
183 # sum the doses (with caution due to units, ...)
184
185 # Compute prescriptions summary table.
186 smmry2 = prescriptions \
187 .groupby(by=[prescriptions.DATE.dt.year, 'DRUG']) \
188 .DRUG.count().rename('use')
189 #.DOSE.sum().rename('use')
190 smmry2.index.names = ['DATE', 'ANTIMICROBIAL']
191
192 # Combine both summary tables
193 smmry = smmry1.reset_index().merge(
194 smmry2.reset_index(), how='inner',
195 left_on=['DATE', 'ANTIMICROBIAL'],
196 right_on=['DATE', 'ANTIMICROBIAL']
197 )
198
199 # Show
200 if TERMINAL:
201 print("\nSummary:")
202 print(smmry)
206 smmry
Lets compute the DRI
212 # -------------------------
213 # Compute DRI
214 # -------------------------
215 # Libraries
216 from pyamr.core.dri import DRI
217
218 # Instance
219 obj = DRI(
220 column_resistance='sari',
221 column_usage='use'
222 )
223
224 # Compute overall DRI
225 dri1 = obj.compute(smmry,
226 groupby=['DATE'],
227 return_usage=True)
228
229 # Compute DRI by organism
230 dri2 = obj.compute(smmry,
231 groupby=['DATE', 'MICROORGANISM'],
232 return_usage=True)
233
234
235 if TERMINAL:
236 print("DRI overall:")
237 print(dri1)
238 print("DRI by microorganism:")
239 print(dri2)
243 dri1
247 dri2
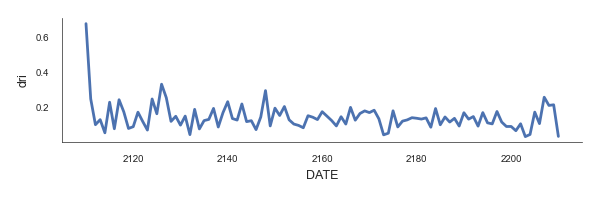
Lets visualise the overall DRI.
252 # --------------------------------------------
253 # Plot
254 # --------------------------------------------
255 # Libraries
256 import matplotlib.pyplot as plt
257 import seaborn as sns
258
259 # Display using relplot
260 g = sns.relplot(data=dri1.reset_index(), x='DATE', y='dri',
261 height=2, aspect=3.0, kind='line',
262 linewidth=2, markersize=0, marker='o'
263 )
264
265 plt.tight_layout()
Lets visualise the microorganism-wise DRI.
270 # --------------------------------------------
271 # Format
272 # --------------------------------------------
273 # Copy results
274 aux = dri2.copy(deep=True)
275
276 # Combine with summary
277 aux = aux.merge(smmry, how='left',
278 left_on=['DATE', 'MICROORGANISM'],
279 right_on=['DATE', 'MICROORGANISM'])
280
281 # Find microorganisms with more samples
282 top = aux.groupby(by='MICROORGANISM') \
283 .freq.sum().sort_values(ascending=False) \
284 .head(4)
285
286 # Filter by top microorganisms
287 aux = aux[aux.MICROORGANISM.isin(top.index)]
288
289 # --------------------------------------------
290 # Plot
291 # --------------------------------------------
292 # Display
293 g = sns.relplot(data=aux,
294 x='DATE', y='dri', hue='MICROORGANISM',
295 row='MICROORGANISM', palette='rocket',
296 #style='event', col='region', palette='palette',
297 height=1.5, aspect=4.0, kind='line', legend=False,
298 linewidth=2, markersize=0, marker='o')
299
300 """
301 # Iterate over each subplot to customize further
302 for title, ax in g.axes_dict.items():
303 ax.text(1., .85, title, transform=ax.transAxes,
304 fontsize=9, fontweight="normal",
305 horizontalalignment='right')
306 """
307 # Configure
308 g.tight_layout()
309 g.set_titles("{row_name}")
310 #g.set_titles("")
311
312 # Show
313 plt.show()
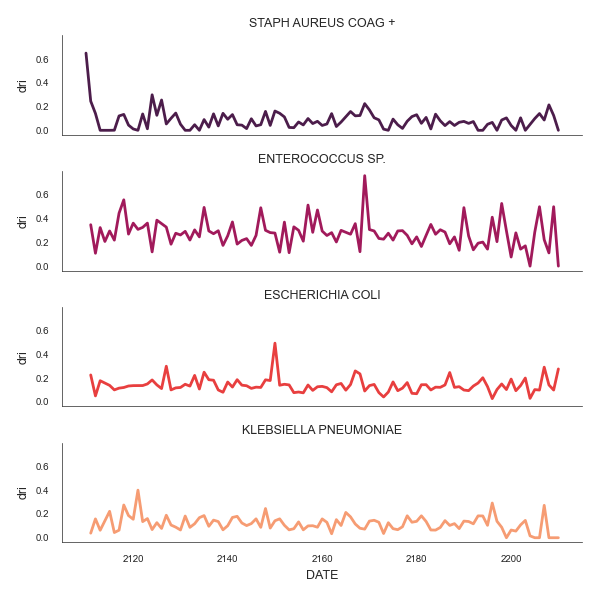
The top microorganisms are:
317 top
MICROORGANISM
ESCHERICHIA COLI 101050.0
STAPH AUREUS COAG + 28165.0
KLEBSIELLA PNEUMONIAE 28094.0
ENTEROCOCCUS SP. 10585.0
Name: freq, dtype: float64
The results look as follows:
321 aux.rename(columns={
322 'intermediate': 'I',
323 'sensitive': 'S',
324 'resistant': 'R',
325 'pass': 'P'
326 }).round(decimals=2)
Total running time of the script: ( 0 minutes 12.707 seconds)