Note
Go to the end to download the full example code
ASAI - Compute timeseries
In order to study the temporal evolution of AMR, it is necessary to generate a resistance time series from the susceptibility test data. This is often achieved by computing the resistance index on consecutive partitions of the data. Note that each partition contains the susceptibility tests required to compute a resistance index. The traditional strategy of dealing with partitions considers independent time intervals (see yearly, monthly or weekly time series in Table 4.2). Unfortunately, this strategy forces to trade-off between granularity (level of detail) and accuracy. On one side, weekly time series are highly granular but inaccurate. On the other hand, yearly time series are accurate but rough. Note that the granularity is represented by the number of observations in a time series while the accuracy is closely related with the number of susceptibility tests used to compute the resistance index. Conversely, the overlapping time intervals strategy drops such dependence by defining a window of fixed size which is moved across time. The length of the window is denoted as period and the time step as shift. For instance, three time series obtained using the overlapping time intervals strategy with a monthly shift (1M) and window lengths of 12, 6 and 3 have been presented for the sake of clarity (see 1M12, 1M6 and 1M3 in Table 4.2).
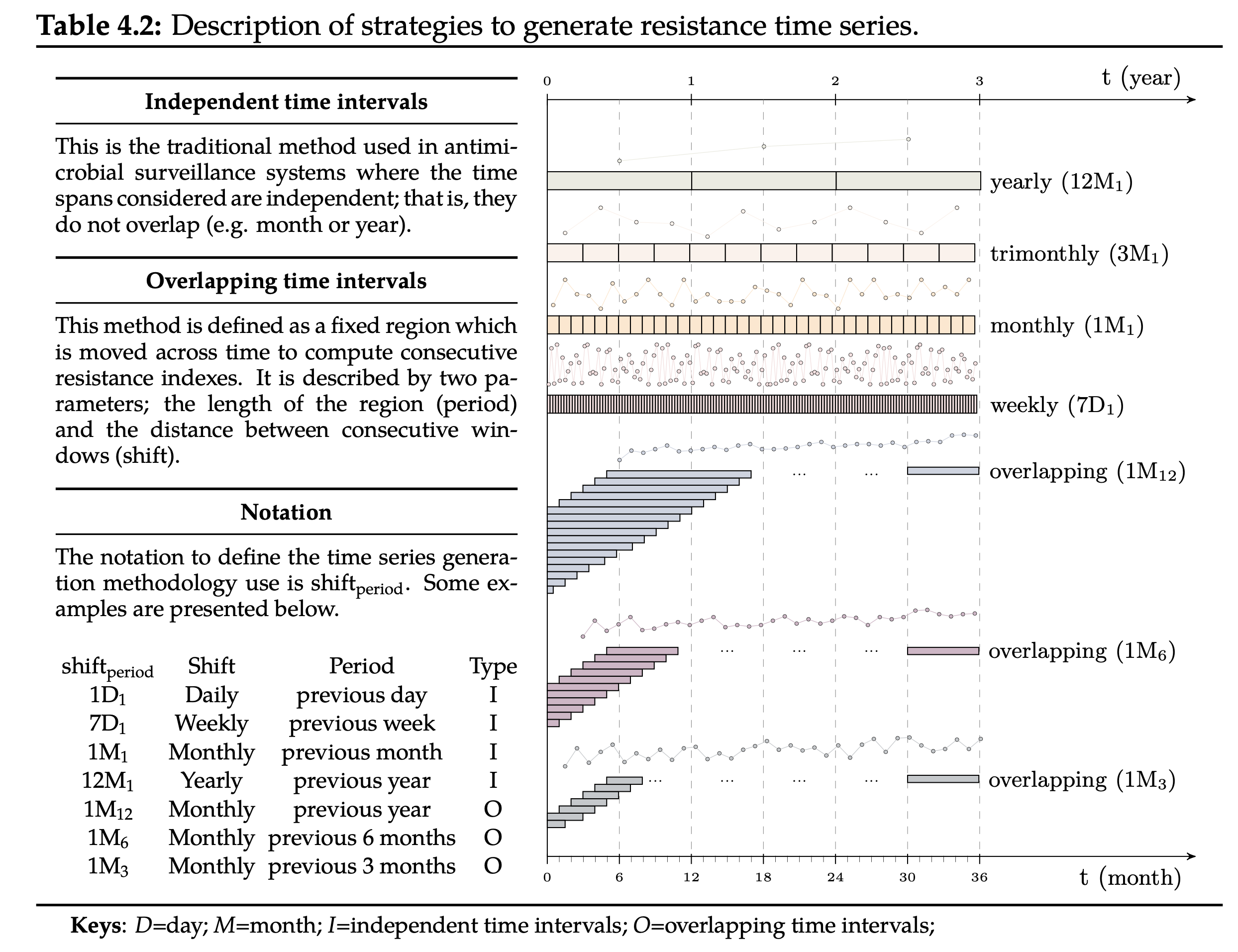
The notation to define the time series generation methodology (SHIFTperiod) is described with various examples in Table 4.2. For instance, 1M12 defines a time series with weekly resistance indexes (7D) calculated using the microbiology records available for the previous four weeks (4x7D). It is important to note that some notations are equivalent representations of the same susceptibility data at different granularities, hence their trends are comparable. As an example, the trend estimated for 1M1 should be approximately thirty times the trend estimated for 1D30.
Let’s see how to compute ASAI time series with examples.
We first load the data and select one single pair for clarity.
54 # Libraries
55 import numpy as np
56 import pandas as pd
57 import seaborn as sns
58 import matplotlib as mpl
59 import matplotlib.pyplot as plt
60
61 # Import own libraries
62 from pyamr.core.sari import sari
63 from pyamr.core.asai import asai
64 from pyamr.datasets.load import load_data_nhs
65
66
67 # -------------------------
68 # Methods
69 # -------------------------
70 def create_mapper(dataframe, column_key, column_value):
71 """This method constructs a mapper
72
73 Parameters
74 ----------
75 dataframe: dataframe-like
76 The dataframe from which the columns are extracted
77
78 column_key: string-like
79 The name of the column with the values for the keys of the mapper
80
81 column_value: string-like
82 The name of the column with the values for the values of the mapper
83
84 Returns
85 -------
86 dictionary
87 """
88 dataframe = dataframe[[column_key, column_value]]
89 dataframe = dataframe.drop_duplicates()
90 return dict(zip(dataframe[column_key], dataframe[column_value]))
91
92 # -------------------------
93 # Configuration
94 # -------------------------
95 # Configure seaborn style (context=talk)
96 sns.set(style="white")
97
98 # Set matplotlib
99 mpl.rcParams['xtick.labelsize'] = 9
100 mpl.rcParams['ytick.labelsize'] = 9
101 mpl.rcParams['axes.titlesize'] = 11
102 mpl.rcParams['legend.fontsize'] = 9
103
104 # Pandas configuration
105 pd.set_option('display.max_colwidth', 40)
106 pd.set_option('display.width', 300)
107 pd.set_option('display.precision', 4)
108
109 # Numpy configuration
110 np.set_printoptions(precision=2)
111
112 # -------------------------------------------
113 # Load data
114 # -------------------------------------------
115 # Load data
116 data, antimicrobials, microorganisms = load_data_nhs()
117
118 # Show
119 print("\nData:")
120 print(data)
121 print("\nColumns:")
122 print(data.columns)
123 print("\nDtypes:")
124 print(data.dtypes)
125
126 # Filter
127 idxs_spec = data.specimen_code.isin(['URICUL'])
128 idxs_abxs = data.antimicrobial_code.isin(['AAUG'])
129
130 # Filter
131 data = data[idxs_spec & idxs_abxs]
132
133 # Filter dates (2016-2018 missing)
134 data = data[data.date_received.between('2008-01-01', '2016-12-31')]
Data:
date_received date_outcome patient_id laboratory_number specimen_code specimen_name ... antimicrobial_code antimicrobial_name sensitivity_method sensitivity mic reported
0 2009-01-03 00:00:00 NaN 20091 X428501 BLDCUL NaN ... AAMI amikacin NaN sensitive NaN NaN
1 2009-01-03 00:00:00 NaN 20091 X428501 BLDCUL NaN ... AAMO amoxycillin NaN resistant NaN NaN
2 2009-01-03 00:00:00 NaN 20091 X428501 BLDCUL NaN ... AAUG augmentin NaN sensitive NaN NaN
3 2009-01-03 00:00:00 NaN 20091 X428501 BLDCUL NaN ... AAZT aztreonam NaN sensitive NaN NaN
4 2009-01-03 00:00:00 NaN 20091 X428501 BLDCUL NaN ... ACAZ ceftazidime NaN sensitive NaN NaN
... ... ... ... ... ... ... ... ... ... ... ... ... ...
7929 2021-01-21 23:56:00 2021-01-22 00:00:00 199863 H2230229 FBCUL Fluid in Blood Culture Bottles ... AGEN gentamicin DD resistant NaN Y
7930 2021-01-21 23:56:00 2021-01-22 00:00:00 199863 H2230229 FBCUL Fluid in Blood Culture Bottles ... AMER meropenem DD sensitive NaN Y
7931 2021-01-21 23:56:00 2021-01-22 00:00:00 199863 H2230229 FBCUL Fluid in Blood Culture Bottles ... ATAZ piperacillin-tazobactam DD sensitive NaN N
7932 2021-01-21 23:56:00 2021-01-22 00:00:00 199863 H2230229 FBCUL Fluid in Blood Culture Bottles ... ATEM temocillin DD sensitive NaN N
7933 2021-01-21 23:56:00 2021-01-22 00:00:00 199863 H2230229 FBCUL Fluid in Blood Culture Bottles ... ATIG tigecycline DD sensitive NaN N
[3770034 rows x 15 columns]
Columns:
Index(['date_received', 'date_outcome', 'patient_id', 'laboratory_number', 'specimen_code', 'specimen_name', 'specimen_description', 'microorganism_code', 'microorganism_name', 'antimicrobial_code', 'antimicrobial_name', 'sensitivity_method', 'sensitivity', 'mic', 'reported'], dtype='object')
Dtypes:
date_received datetime64[ns]
date_outcome object
patient_id object
laboratory_number object
specimen_code object
specimen_name object
specimen_description object
microorganism_code object
microorganism_name object
antimicrobial_code object
antimicrobial_name object
sensitivity_method object
sensitivity object
mic object
reported object
dtype: object
Independent Time Intervals (ITI)
This is the traditional method used in antimicrobial surveillance systems where the time spans considered are independent; that is, they do not overlap (e.g. monthly time series - 1M1 or yearly timeseries - 12M1).
Todo
Include in ASAI an option to filter and keep only those genus and species that have values over all the time period? or at least for more than 80 percent of the period?
150 # -------------------------------------------
151 # Compute ITI sari (temporal)
152 # -------------------------------------------
153 from pyamr.core.sari import SARI
154
155 # Create SARI instance
156 sari = SARI(groupby=['specimen_code',
157 'microorganism_code',
158 'antimicrobial_code',
159 'sensitivity'])
160
161 # Create constants
162 shift, period = '6M', '800D'
163
164 # Compute sari timeseries
165 temp = sari.compute(data, shift=shift,
166 period=period, cdate='date_received')
167
168 # Reset index
169 temp = temp.reset_index()
170
171 # Show
172 print("\nSARI (temporal)")
173 print(temp)
174
175
176 # -------------------------------------------
177 # Filter for sari (temporal)
178 # -------------------------------------------
179 # Appearance of organisms by date (or even freq instead of 1/0).
180 s = pd.crosstab(temp['microorganism_code'], temp['date_received'])
181
182 # Filter
183 #temp = temp[temp['microorganism_code'].isin(s[s.all(axis=1)].index)]
184
185 # Show
186 print("Remaining microorganisms:")
187 print(temp.microorganism_code.unique())
188
189 # -------------------------
190 # Format dataframe
191 # -------------------------
192 # Create mappers
193 abx_map = create_mapper(antimicrobials, 'antimicrobial_code', 'category')
194 org_map = create_mapper(microorganisms, 'microorganism_code', 'genus')
195 grm_map = create_mapper(microorganisms, 'microorganism_code', 'gram_stain')
196
197 #iti = iti[['date_received', 'freq', 'sari', 'antimicrobial_code', 'microorganism_code']]
198
199 # Include categories
200 temp['category'] = temp['antimicrobial_code'].map(abx_map)
201 temp['genus'] = temp['microorganism_code'].map(org_map)
202 temp['gram'] = temp['microorganism_code'].map(grm_map)
203
204 # Empty grams are a new category (unknown - u)
205 temp.gram = temp.gram.fillna('u')
206
207 # Show
208 print("\nMicroorganisms without gram stain:")
209 print(temp[temp.gram=='u'].microorganism_code.unique())
210
211 # -------------------------------
212 # Create antimicrobial spectrum
213 # -------------------------------
214 # Libraries
215 from pyamr.core.asai import ASAI
216
217 # Create antimicrobial spectrum of activity instance
218 asai = ASAI(column_genus='genus',
219 column_specie='microorganism_code',
220 column_resistance='sari',
221 column_frequency='freq')
222 # Compute
223 scores = asai.compute(temp,
224 groupby=['date_received',
225 'antimicrobial_code',
226 'gram'],
227 weights='uniform',
228 threshold=None,
229 min_freq=0)
230
231 # Unstack
232 scores = scores
233
234 # Show scores
235 print("\nASAI:")
236 print(scores.unstack())
237
238 # Plot
239 # ----
240 # Create figure
241 fig, axes = plt.subplots(1, 2, figsize=(10, 3), sharey=True)
242
243 # Show
244 sns.lineplot(data=scores, x='date_received', y='ASAI_SCORE',
245 hue='gram', palette="tab10", linewidth=0.75, linestyle='--',
246 marker='o', markersize=3, markeredgecolor='k',
247 markeredgewidth=0.5, markerfacecolor=None,
248 alpha=0.5, ax=axes[0])
249
250
251 plt.show()
252
253 """
254 # -------------------------
255 # Compute ASAI
256 # -------------------------
257 iti = iti.rename(columns={
258 'microorganism_code': 'SPECIE',
259 'sari': 'RESISTANCE',
260 'genus': 'GENUS'
261 })
262
263
264 fig, axes = plt.subplots(1, 2, figsize=(10, 3), sharey=True)
265
266 # Variable for filtering
267 s = pd.crosstab(iti['SPECIE'], iti['date_received'])
268
269 # Create aux (non filtered)
270 aux = iti.copy(deep=True)
271 aux = aux.groupby(['date_received',
272 'antimicrobial_code',
273 'gram'])\
274 .apply(asai, weights='uniform', threshold=0.5)
275
276
277 plt.show()
278
279 # Create aux (filtered)
280 aux2 = iti.copy(deep=True)
281 aux2 = aux2[aux2['SPECIE'].isin(s[s.all(axis=1)].index)]
282 aux2 = aux2.groupby(['date_received',
283 'antimicrobial_code',
284 'gram'])\
285 .apply(asai, weights='uniform', threshold=0.8)
286
287 a = 1
288 # Display
289 print("\nCrosstab:")
290 print(s)
291 print("\nSelected:")
292 print(s[s.all(axis=1)])
293 print("\nRemaining:")
294 print(iti)
295 print(aux.unstack())
296 print(aux2.unstack())
297
298 sns.lineplot(data=aux.reset_index(),
299 x='date_received',
300 y='ASAI_SCORE',
301 palette="tab10",
302 linewidth=0.75,
303 hue='gram',
304 marker='o',
305 ax=axes[0])
306
307 sns.lineplot(data=aux2.reset_index(),
308 x='date_received',
309 y='ASAI_SCORE',
310 palette="tab10",
311 linewidth=0.75,
312 hue='gram',
313 marker='o',
314 ax=axes[1])
315 """
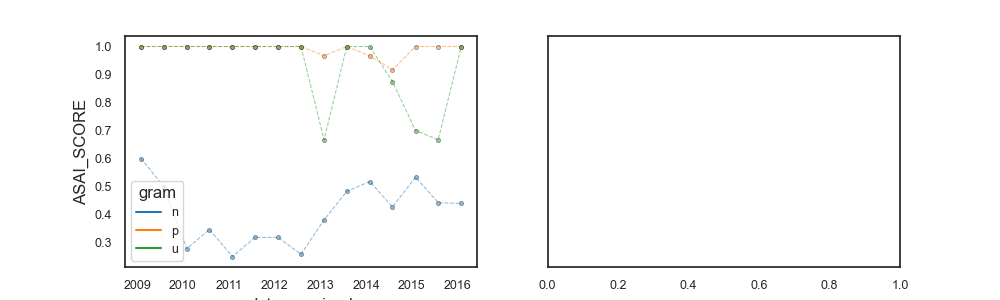
SARI (temporal)
specimen_code microorganism_code antimicrobial_code date_received hide intermediate not done resistant sensitive freq sari
0 URICUL ABAU AAUG 2009-07-31 0.0 0.0 0.0 3.0 2.0 5.0 0.6000
1 URICUL ABAU AAUG 2010-01-31 0.0 0.0 0.0 4.0 3.0 7.0 0.5714
2 URICUL ABAU AAUG 2010-07-31 0.0 0.0 0.0 8.0 6.0 14.0 0.5714
3 URICUL ABAU AAUG 2011-01-31 0.0 0.0 0.0 11.0 8.0 19.0 0.5789
4 URICUL ABAU AAUG 2011-07-31 0.0 0.0 0.0 13.0 9.0 22.0 0.5909
.. ... ... ... ... ... ... ... ... ... ... ...
514 URICUL VIRST AAUG 2012-07-31 0.0 0.0 0.0 0.0 1.0 1.0 0.0000
515 URICUL VIRST AAUG 2013-07-31 0.0 0.0 0.0 0.0 2.0 2.0 0.0000
516 URICUL VIRST AAUG 2014-01-31 0.0 0.0 0.0 0.0 3.0 3.0 0.0000
517 URICUL YEAST AAUG 2015-01-31 0.0 0.0 0.0 0.0 1.0 1.0 0.0000
518 URICUL YEAST AAUG 2015-07-31 0.0 0.0 0.0 0.0 2.0 2.0 0.0000
[519 rows x 11 columns]
Remaining microorganisms:
['ABAU' 'ACHRO' 'ACINE' 'AEROC' 'AEROM' 'AFAE' 'AHS' 'AHYD' 'ALWO' 'AVIR'
'AXYL' 'A_ARADIORES' 'A_BSGD' 'A_CHRYSEOB' 'A_CHRYSEOM' 'A_CTESTOSTE'
'A_EAVIUM' 'A_ELUDWIGII' 'A_FLAVOBAC' 'A_HAFNIA S' 'A_ISOLATED'
'A_LACTOBAC' 'A_MBG' 'A_PROVIDEN' 'A_RAOULTEL' 'A_SBOVIS' 'A_SODORIFER'
'BACIL' 'BCEP' 'BHSA' 'BHSB' 'BHSC' 'BHSCG' 'BHSG' 'CAMA' 'CBRA' 'CFRE'
'CIND' 'CITRO' 'CKOS' 'CNS' 'COLIF' 'CORYN' 'CSTR' 'DACI' 'DIPHT' 'EAER'
'EASB' 'ECLO' 'ECOL' 'EFAM' 'EFAS' 'ENTB' 'ENTC' 'HAEMO' 'HALV' 'HINF'
'KLEBS' 'KOXY' 'KPNE' 'LFC' 'MCOL' 'MICROC' 'MMOR' 'MODO' 'NEISS' 'NLF'
'PAER' 'PANTO' 'PMIR' 'PPUT' 'PRET' 'PROTE' 'PSEUD' 'PSTUA' 'PVUL'
'QMCOL' 'QMIXY' 'SAGA' 'SALMO' 'SAUR' 'SEPI' 'SERRA' 'SHAEM' 'SLIQ'
'SLUG' 'SMAL' 'SMAR' 'SMIL' 'SMIT' 'SPYO' 'SSAL' 'SSAP' 'STAPH' 'STREP'
'VIRST' 'YEAST']
Microorganisms without gram stain:
['A_CHRYSEOM' 'A_ISOLATED' 'A_MBG' 'COLIF' 'DIPHT' 'LFC' 'MCOL' 'MODO'
'NLF' 'QMCOL' 'QMIXY' 'YEAST']
ASAI:
N_GENUS N_SPECIE ASAI_SCORE
gram n p u n p u n p u
date_received antimicrobial_code
2009-01-31 AAUG 5.0 3.0 1.0 5.0 4.0 1.0 0.6000 1.0000 1.0000
2009-07-31 AAUG 13.0 3.0 1.0 14.0 8.0 1.0 0.5000 1.0000 1.0000
2010-01-31 AAUG 9.0 3.0 1.0 11.0 7.0 1.0 0.2778 1.0000 1.0000
2010-07-31 AAUG 13.0 3.0 2.0 15.0 8.0 2.0 0.3462 1.0000 1.0000
2011-01-31 AAUG 10.0 4.0 1.0 11.0 10.0 1.0 0.2500 1.0000 1.0000
2011-07-31 AAUG 11.0 3.0 1.0 12.0 6.0 1.0 0.3182 1.0000 1.0000
2012-01-31 AAUG 11.0 4.0 1.0 14.0 7.0 1.0 0.3182 1.0000 1.0000
2012-07-31 AAUG 11.0 4.0 2.0 21.0 10.0 3.0 0.2576 1.0000 1.0000
2013-01-31 AAUG 14.0 6.0 3.0 29.0 17.0 5.0 0.3810 0.9667 0.6667
2013-07-31 AAUG 19.0 4.0 3.0 36.0 13.0 3.0 0.4825 1.0000 1.0000
2014-01-31 AAUG 14.0 6.0 4.0 34.0 14.0 5.0 0.5179 0.9667 1.0000
2014-07-31 AAUG 16.0 4.0 4.0 36.0 16.0 6.0 0.4271 0.9167 0.8750
2015-01-31 AAUG 16.0 6.0 5.0 34.0 19.0 7.0 0.5333 1.0000 0.7000
2015-07-31 AAUG 13.0 4.0 3.0 27.0 13.0 3.0 0.4423 1.0000 0.6667
2016-01-31 AAUG 11.0 1.0 2.0 22.0 4.0 2.0 0.4394 1.0000 1.0000
'\n# -------------------------\n# Compute ASAI\n# -------------------------\niti = iti.rename(columns={\n \'microorganism_code\': \'SPECIE\',\n \'sari\': \'RESISTANCE\',\n \'genus\': \'GENUS\'\n})\n\n\nfig, axes = plt.subplots(1, 2, figsize=(10, 3), sharey=True)\n\n# Variable for filtering\ns = pd.crosstab(iti[\'SPECIE\'], iti[\'date_received\'])\n\n# Create aux (non filtered)\naux = iti.copy(deep=True)\naux = aux.groupby([\'date_received\',\n \'antimicrobial_code\',\n \'gram\']) .apply(asai, weights=\'uniform\', threshold=0.5)\n\n\nplt.show()\n\n# Create aux (filtered)\naux2 = iti.copy(deep=True)\naux2 = aux2[aux2[\'SPECIE\'].isin(s[s.all(axis=1)].index)]\naux2 = aux2.groupby([\'date_received\',\n \'antimicrobial_code\',\n \'gram\']) .apply(asai, weights=\'uniform\', threshold=0.8)\n\na = 1\n# Display\nprint("\nCrosstab:")\nprint(s)\nprint("\nSelected:")\nprint(s[s.all(axis=1)])\nprint("\nRemaining:")\nprint(iti)\nprint(aux.unstack())\nprint(aux2.unstack())\n\nsns.lineplot(data=aux.reset_index(),\n x=\'date_received\',\n y=\'ASAI_SCORE\',\n palette="tab10",\n linewidth=0.75,\n hue=\'gram\',\n marker=\'o\',\n ax=axes[0])\n\nsns.lineplot(data=aux2.reset_index(),\n x=\'date_received\',\n y=\'ASAI_SCORE\',\n palette="tab10",\n linewidth=0.75,\n hue=\'gram\',\n marker=\'o\',\n ax=axes[1])\n'
Total running time of the script: ( 0 minutes 13.933 seconds)