Note
Go to the end to download the full example code
ACSI - Example using MIMIC
7 # Libraries
8 import sys
9 import warnings
10 import numpy as np
11 import pandas as pd
12 import seaborn as sns
13 import matplotlib as mpl
14
15 from pathlib import Path
16
17 try:
18 __file__
19 TERMINAL = True
20 except:
21 TERMINAL = False
22
23
24 # Configure seaborn style (context=talk)
25 sns.set_theme(style="white")
26
27 # Configure warnings
28 warnings.filterwarnings("ignore",
29 category=pd.errors.DtypeWarning)
30
31 # -------------------------------------------------------
32 # Constants
33 # -------------------------------------------------------
34 # Rename columns for susceptibility
35 rename_susceptibility = {
36 'chartdate': 'DATE',
37 'micro_specimen_id': 'LAB_NUMBER',
38 'spec_type_desc': 'SPECIMEN',
39 'org_name': 'MICROORGANISM',
40 'ab_name': 'ANTIMICROBIAL',
41 'interpretation': 'SENSITIVITY'
42 }
Let’s load the susceptibility test data
48 # -----------------------------
49 # Load susceptibility test data
50 # -----------------------------
51 nrows=1000
52
53 # Helper
54 subset = rename_susceptibility.values()
55
56 # Load data
57 path = Path('../../pyamr/datasets/mimic')
58 data1 = pd.read_csv(path / 'susceptibility.csv', nrows=nrows)
59
60 # Rename columns
61 data1 = data1.rename(columns=rename_susceptibility)
62
63 # Format data
64 data1 = data1[subset]
65 data1 = data1.dropna(subset=subset, how='any')
66 data1.DATE = pd.to_datetime(data1.DATE)
69 data1.head(5)
Let’s compute the ACSI and return the combinations.
Note
This step is quite computationally expensive since it has to create all possible antimicrobial combinations within each isolate in the susceptibility test data. Thus, it is recommended to save the results for future analysis and/or visualisation.
80 # Libraries
81 from pyamr.core.acsi import ACSI
82
83 # Compute index
84 contingency, combinations = \
85 ACSI().compute(data1,
86 groupby=[
87 'DATE',
88 'SPECIMEN',
89 'MICROORGANISM'
90 ],
91 return_combinations=True)
92
93 # Save
94 #contingency.to_csv('contingency_%s.csv' % nrows)
95 #combinations.to_csv('combinations_%s.csv' % nrows)
96
97 # Display
98 print("\nCombinations:")
99 print(combinations)
100 print("\nContingency:")
101 print(contingency)
c:\users\kelda\desktop\repositories\github\pyamr\main\pyamr\core\acsi.py:120: RuntimeWarning:
invalid value encountered in divide
c:\users\kelda\desktop\repositories\github\pyamr\main\pyamr\core\acsi.py:121: RuntimeWarning:
invalid value encountered in divide
Combinations:
DATE SPECIMEN MICROORGANISM LAB_NUMBER ANTIMICROBIAL_x ANTIMICROBIAL_y SENSITIVITY_x SENSITIVITY_y class
0 2112-10-05 BLOOD CULTURE VIRIDANS S... 3530668 CLINDAMYCIN ERYTHROMYCIN S R SR
1 2112-10-05 BLOOD CULTURE VIRIDANS S... 3530668 CLINDAMYCIN PENICILLIN G S S SS
2 2112-10-05 BLOOD CULTURE VIRIDANS S... 3530668 CLINDAMYCIN VANCOMYCIN S S SS
3 2112-10-05 BLOOD CULTURE VIRIDANS S... 3530668 ERYTHROMYCIN PENICILLIN G R S RS
4 2112-10-05 BLOOD CULTURE VIRIDANS S... 3530668 ERYTHROMYCIN VANCOMYCIN R S RS
... ... ... ... ... ... ... ... ... ...
5074 2202-04-26 URINE MORGANELLA... 8654517 NITROFURAN... TOBRAMYCIN R R RR
5075 2202-04-26 URINE MORGANELLA... 8654517 NITROFURAN... TRIMETHOPR... R R RR
5076 2202-04-26 URINE MORGANELLA... 8654517 PIPERACILL... TOBRAMYCIN S R SR
5077 2202-04-26 URINE MORGANELLA... 8654517 PIPERACILL... TRIMETHOPR... S R SR
5078 2202-04-26 URINE MORGANELLA... 8654517 TOBRAMYCIN TRIMETHOPR... R R RR
[5079 rows x 9 columns]
Contingency:
class II IR IS RI RR RS SI SR SS acsi
DATE SPECIMEN MICROORGANISM ANTIMICROBI... ANTIMICROBI...
2112-10-05 BLOOD CULTURE VIRIDANS ST... CLINDAMYCIN ERYTHROMYCIN NaN NaN NaN NaN NaN NaN NaN 1.0 NaN 0.0
PENICILLIN G NaN NaN NaN NaN NaN NaN NaN NaN 1.0 0.0
VANCOMYCIN NaN NaN NaN NaN NaN NaN NaN NaN 1.0 0.0
ERYTHROMYCIN PENICILLIN G NaN NaN NaN NaN NaN 1.0 NaN NaN NaN 0.0
VANCOMYCIN NaN NaN NaN NaN NaN 1.0 NaN NaN NaN 0.0
... .. .. .. .. ... ... .. ... ... ...
2202-04-26 URINE MORGANELLA ... NITROFURANTOIN TOBRAMYCIN NaN NaN NaN NaN 1.0 NaN NaN NaN NaN 0.0
TRIMETHOPRI... NaN NaN NaN NaN 1.0 NaN NaN NaN NaN 0.0
PIPERACILLI... TOBRAMYCIN NaN NaN NaN NaN NaN NaN NaN 1.0 NaN 0.0
TRIMETHOPRI... NaN NaN NaN NaN NaN NaN NaN 1.0 NaN 0.0
TOBRAMYCIN TRIMETHOPRI... NaN NaN NaN NaN 1.0 NaN NaN NaN NaN 0.0
[4464 rows x 10 columns]
Let’s compute the ACSI.
Note
We are loading previously computed combinations.
112 # Libraries
113 from pyamr.datasets.load import fixture
114
115 # Path
116 combinations = fixture(name='mimic/asci/combinations.csv')
117
118 # Lets compute the overall index
119 contingency = ACSI().compute(
120 combinations.reset_index(),
121 groupby=[],
122 flag_combinations=True,
123 return_combinations=False
124 )
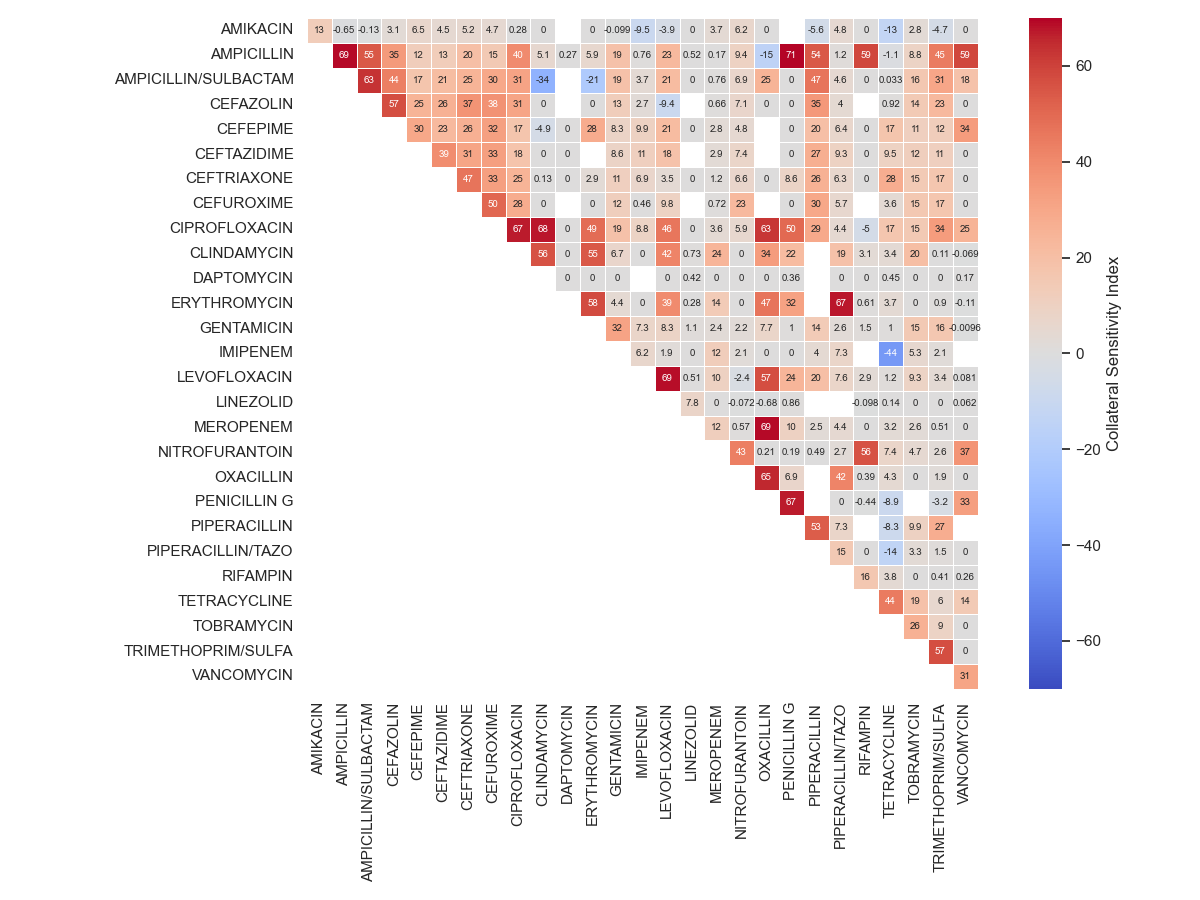
Let’s visualise the result
130 # ------------------------------------------
131 # Display
132 # ------------------------------------------
133 # Display
134 import numpy as np
135 import seaborn as sns
136 import matplotlib.pyplot as plt
137
138 # Get unique antimicrobials
139 s1 = set(combinations.ANTIMICROBIAL_x.unique())
140 s2 = set(combinations.ANTIMICROBIAL_x.unique())
141 abxs = s1.union(s2)
142
143 # Create index with all pairs
144 index = pd.MultiIndex.from_product([abxs, abxs])
145
146 # Reformat
147 aux = contingency['acsi'] \
148 .reindex(index, fill_value=np.nan) \
149 .unstack()
150
151 # Create figure
152 fig, axs = plt.subplots(nrows=1, ncols=1,
153 sharey=False, sharex=False, figsize=(12, 9)
154 )
155
156 # Display
157 sns.heatmap(data=aux * 100, ax=axs,
158 annot=True, annot_kws={'size':7}, square=True,
159 linewidth=.5, xticklabels=True, yticklabels=True,
160 cmap='coolwarm', vmin=-70, vmax=70, center=0,
161 cbar_kws={'label': 'Collateral Sensitivity Index'})
162
163 # Show
164 plt.tight_layout()
165 plt.show()
Total running time of the script: ( 0 minutes 7.821 seconds)