Note
Go to the end to download the full example code
Table Boxplot
Data:
org abx sari sart r2 f_prob jb jb_prob ptm ptn dw surveillance
0 ABAU AAMI 0.3906 -5.0000e-03 0.181 2.9400e-05 4.648 9.7900e-02 0.000 0.000 1.177 M
1 ABAU AAMO 0.9603 -1.2000e-03 0.053 1.9400e-02 631.965 5.9000e-138 0.019 0.000 1.848 M
2 ABAU AAUG 0.7578 -8.0000e-04 -0.004 4.2400e-01 13.083 1.4400e-03 0.424 0.000 1.640 M
3 ABAU AAZT 0.9706 -9.0000e-04 0.057 1.5600e-02 129.263 8.5300e-29 0.016 0.000 1.604 M
4 ABAU ACAZ 0.9417 -7.0000e-04 0.015 1.3200e-01 103.616 3.1600e-23 0.132 0.000 1.776 M
... ... ... ... ... ... ... ... ... ... ... ... ...
1726 SSAP AGEN 0.0026 -6.3420e-05 0.027 7.2100e-02 40.688 1.4600e-09 0.072 0.004 0.180 SW
1727 SSAP AMET 0.2897 3.8000e-03 0.539 5.1000e-16 2.913 2.3300e-01 0.000 0.000 0.115 SW
1728 SSAP ANIT 0.0118 -2.0000e-04 0.418 1.0700e-11 3.205 2.0100e-01 0.000 0.000 0.443 SW
1729 SSAP APEN 0.8494 -5.0000e-04 0.042 3.2400e-02 5.323 6.9800e-02 0.032 0.000 0.308 SW
1730 SSAP ATRI 0.0608 -1.6810e-05 -0.011 8.0200e-01 0.007 9.9700e-01 0.802 0.000 0.316 SW
[1731 rows x 12 columns]
6 # Libraries
7 import numpy as np
8 import pandas as pd
9 import seaborn as sns
10 import matplotlib.pyplot as plt
11 import matplotlib.gridspec as gridspec
12
13 from matplotlib import colors
14
15 from pyamr.utils.plot import hlinebgplot
16
17 import pyamr
18 pyamr.utils.plot.scalar_colormap([1,2,3,4], 'coolwarm', vmin=0, vmax=10)
19
20 # -------------------
21 # PLOTTING SETTINGS
22 # -------------------
23 # Configuration for each columns
24 info_sari = {
25 'cmap':'Reds',
26 'title':'SARI',
27 'xlim':[-0.1, 1.1],
28 'xticks':[0, 1],
29 }
30
31 info_sart = {
32 'cmap':'RdBu_r',
33 'title':'SART',
34 'xlim':[-1.2, 1.2],
35 'xticks':[-1, 1],
36 }
37
38 info_r2 = {
39 'cmap':'YlGn',
40 'title':'R2',
41 'ylim': [-0.2, 1.2],
42 'yticks': [.0, .2, .4, .6, .8 ]
43 }
44
45 info_fprob = {
46 'name':'f_prob',
47 'title':'Prob(F)',
48 'ylim': [-0.2, 1.2],
49 'yticks': [.0, .2, .4, .6, .8 ]
50 }
51
52 info_jb_prob = {
53 'cmap':'YlGn',
54 'title':'Prob(JB)',
55 'ylim': [-0.2, 1.2],
56 'hline': [{'yv':0.05, 'bg':0.0}],
57 'yticks': [.0, .2, .4, .6, .8 ]
58 }
59
60 info_dw = {
61 'cmap':'YlGn_r',
62 'title':'DW',
63 'hline': [{'yv':0.8, 'bg':4.0}],
64 'ylim': [-0, 4.2],
65 'yticks': [1, 2, 3]
66 }
67
68 info_slope = {
69 'name':'x1_tprob',
70 'cmap':'YlGn_r',
71 'title':'P>|t| m',
72 'hline': [{'yv':0.05, 'bg':1.0}],
73 'ylim': [-0.2, 1.2],
74 'yticks': [.0, .2, .4, .6, .8 ]
75 }
76
77 info_coefficient = {
78 'name':'c_tprob',
79 'cmap':'YlGn_r',
80 'title':'P>|t| n',
81 'hline': [{'yv':0.05, 'bg':1.0}],
82 'ylim': [-0.2, 1.2],
83 'yticks': [.0, .2, .4, .6, .8 ]
84 }
85
86
87 # Now we combine all of them together. Note
88 # that the key value corresponds to the name
89 # of the column it should be applied to.
90 info = {
91 'sari': info_sari,
92 'sart': info_sart,
93 'dw': info_dw,
94 'r2': info_r2,
95 'jb_prob': info_jb_prob,
96 'ptm': info_slope,
97 'ptn': info_coefficient,
98 'f_prob': info_fprob
99 }
100
101 # -------------------------------------------
102 # Main
103 # -------------------------------------------
104 # Data path.
105 path = '../../pyamr/fixtures/fixture_surveillance.csv'
106
107 # Rename
108 rename = {
109 'organismCode': 'org',
110 'antibioticCode': 'abx',
111 'sari': 'sari',
112 'x1_coef': 'sart',
113 'adj_rsquared': 'r2',
114 'f_prob': 'f_prob',
115 'jarque-bera': 'jb',
116 'jarque_bera_prob': 'jb_prob',
117 'x1_tprob': 'ptm',
118 'c_tprob': 'ptn',
119 'durbin-watson': 'dw',
120 'surveillance': 'surveillance'
121 }
122
123 # Create data
124 data = pd.read_csv(path)
125 data = data[rename.keys()]
126 data = data.rename(columns=rename)
127
128 # Show
129 print("\nData:")
130 print(data)
131
132
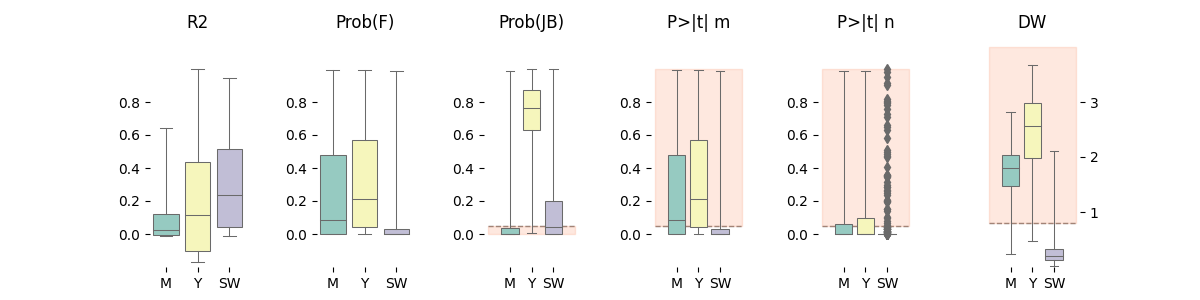
133 # -------------------------------------------
134 # Pair Grid
135 # -------------------------------------------
136 # Features
137 statistics = ['r2', 'f_prob', 'jb_prob', 'ptm', 'ptn', 'dw']
138
139 # Create figure
140 f, axes = plt.subplots(1, len(statistics), figsize=(12, 3))
141 axes = axes.flatten()
142
143 # Plot strips and format axes
144 for ax, c in zip(axes, statistics):
145
146 # Get information
147 d = info[c] if c in info else {}
148
149 # Plot distributions
150 sns.boxplot(x='surveillance', y=data[c],
151 ax=ax, data=data, whis=1e30,
152 palette="Set3", linewidth=0.75)
153
154 # Plot horizontal lines
155 for e in d.get('hline', []):
156 hlinebgplot(ax, right=3, **e)
157
158 # Configure axes
159 ax.set(title=d.get('title', c),
160 xlabel='', ylabel='',
161 ylim=d.get('ylim', None),
162 yticks=d.get('yticks', ax.get_yticks()))
163 #ax.tick_params(axis='y', which='both', length=0)
164 ax.xaxis.grid(False)
165 ax.yaxis.grid(False)
166
167 # Despine
168 sns.despine(bottom=True, left=True)
169
170 # Change last ticks to right
171 axes[-1].yaxis.tick_right()
172
173 # Adjust space (overwritten by tight)
174 plt.subplots_adjust(wspace=0.75)
175
176 # Configure layout
177 #plt.tight_layout()
178
179 # Show
180 plt.show()
Total running time of the script: ( 0 minutes 0.314 seconds)