Note
Go to the end to download the full example code
Step 02 - Temporal evolution
In this example, we will explore how to compute a time series from susceptibility test data and examine different indexes that can be utilized. Through this analysis, we will gain a deeper understanding of how to utilize these indexes to evaluate the evolving patterns of bacterial susceptibility and guide effective antimicrobial therapy strategies.
In order to study the temporal evolution of AMR, it is necessary to generate a resistance time series from the susceptibility test data. This is often achieved by computing the resistance index on consecutive partitions of the data. Note that each partition contains the susceptibility tests required to compute a resistance index. The traditional strategy of dealing with partitions considers independent time intervals (see yearly, monthly or weekly time series in Table 4.2). Unfortunately, this strategy forces to trade-off between granularity (level of detail) and accuracy. On one side, weekly time series are highly granular but inaccurate. On the other hand, yearly time series are accurate but rough. Note that the granularity is represented by the number of observations in a time series while the accuracy is closely related with the number of susceptibility tests used to compute the resistance index. Conversely, the overlapping time intervals strategy drops such dependence by defining a window of fixed size which is moved across time. The length of the window is denoted as period and the time step as shift. For instance, three time series obtained using the overlapping time intervals strategy with a monthly shift (1M) and window lengths of 12, 6 and 3 have been presented for the sake of clarity (see 1M12, 1M6 and 1M3 in Table 4.2).
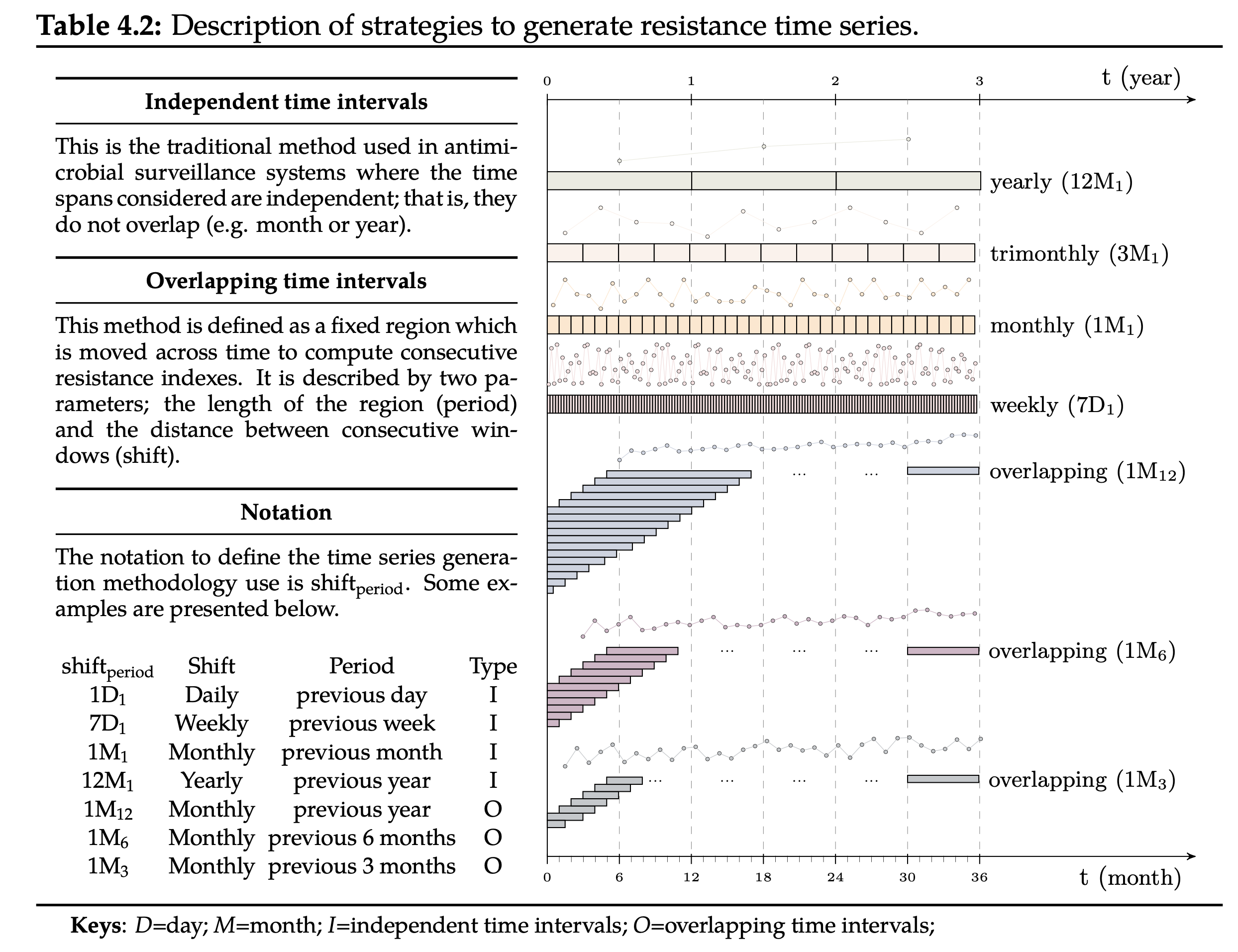
The notation to define the time series generation methodology (SHIFTperiod) is described with various examples in Table 4.2. For instance, 1M12 defines a time series with weekly resistance indexes (7D) calculated using the microbiology records available for the previous four weeks (4x7D). It is important to note that some notations are equivalent representations of the same susceptibility data at different granularities, hence their trends are comparable. As an example, the trend estimated for 1M1 should be approximately thirty times the trend estimated for 1D30.
Note
Using overlapping time intervals to compute an index is better than applying a moving average because it captures more detailed patterns in the data and reduces bias from fixed window sizes. It provides a more comprehensive analysis and improves accuracy in representing the characteristics of the time series.
Loading data
A small dataset will be used for this example.
65 # Libraries
66 import numpy as np
67 import pandas as pd
68 import seaborn as sns
69 import matplotlib as mpl
70 import matplotlib.pyplot as plt
71
72 # Import from pyAMR
73 from pyamr.datasets.load import make_susceptibility
74
75 # -------------------------------------------
76 # Load data
77 # -------------------------------------------
78 # Load data
79 data = make_susceptibility()
80 data = data.drop_duplicates()
81
82 # Convert date to datetime
83 data.date_received = pd.to_datetime(data.date_received)
84
85 # Filter (speeds up the execution)
86 idxs_spec = data.specimen_code.isin(['URICUL'])
87 idxs_abxs = data.antimicrobial_name.isin(['augmentin'])
88
89 # Filter
90 data = data[idxs_spec & idxs_abxs]
91
92 # Show
93 print("\nData:")
94 print(data)
95 print("\nColumns:")
96 print(data.dtypes)
Data:
date_received date_outcome patient_id laboratory_number specimen_code specimen_name specimen_description ... microorganism_name antimicrobial_code antimicrobial_name sensitivity_method sensitivity mic reported
113 2009-01-03 NaN 20099 X428892 URICUL NaN urine ... coliform AAUG augmentin NaN sensitive NaN NaN
119 2009-01-03 NaN 20100 X429141 URICUL NaN catheter urine ... coliform AAUG augmentin NaN sensitive NaN NaN
164 2009-01-03 NaN 22571 X429323 URICUL NaN mid stream urine ... coliform AAUG augmentin NaN sensitive NaN NaN
268 2009-01-03 NaN 22576 X428467 URICUL NaN mid stream urine ... escherichia coli AAUG augmentin NaN sensitive NaN NaN
293 2009-01-03 NaN 24017 X429325 URICUL NaN clean catch urine ... escherichia coli AAUG augmentin NaN sensitive NaN NaN
... ... ... ... ... ... ... ... ... ... ... ... ... ... .. ...
318940 2009-12-31 NaN 20088 H2011867 URICUL NaN mid stream urine ... escherichia coli AAUG augmentin NaN sensitive NaN NaN
318946 2009-12-31 NaN 20089 H2012653 URICUL NaN mid stream urine ... escherichia coli AAUG augmentin NaN sensitive NaN NaN
318983 2009-12-31 NaN 22565 F1741389 URICUL NaN mid stream urine ... escherichia coli AAUG augmentin NaN sensitive NaN NaN
319048 2009-12-31 NaN 24013 H2012150 URICUL NaN urine ... enterococcus AAUG augmentin NaN sensitive NaN NaN
319070 2009-12-31 NaN 24015 H2012340 URICUL NaN mid stream urine ... escherichia coli AAUG augmentin NaN sensitive NaN NaN
[15269 rows x 15 columns]
Columns:
date_received datetime64[ns]
date_outcome float64
patient_id int64
laboratory_number object
specimen_code object
specimen_name float64
specimen_description object
microorganism_code object
microorganism_name object
antimicrobial_code object
antimicrobial_name object
sensitivity_method float64
sensitivity object
mic float64
reported float64
dtype: object
Computing SARI timeseries
In order to study the temporal evolution of AMR, it is necessary to generate
a resistance time series from the susceptibility test data. This is often
achieved by calculating the resistance index; that is SARI on consecutive
partitions of the data. Note that each partition contains the susceptibility
tests that will be used to compute the resistance index.
For more information see: pyamr.core.sari.SARI
For more examples see:
First, let’s compute the time series
116 # -----------------------------------------
117 # Compute sari (temporal)
118 # -----------------------------------------
119 from pyamr.core.sari import SARI
120
121 # Create SARI instance
122 sar = SARI(groupby=['specimen_code',
123 'microorganism_name',
124 'antimicrobial_name',
125 'sensitivity'])
126
127 # Create constants
128 shift, period = '30D', '30D'
129
130 # Compute sari timeseries
131 iti = sar.compute(data, shift=shift,
132 period=period, cdate='date_received')
133
134 # Reset index
135 iti = iti.reset_index()
136
137 # Show
138 #print("\nSARI (temporal):")
139 #print(iti)
140
141 iti.head(10)
Let’s plot the evolution of a single combination.
148 # --------------
149 # Filter
150 # --------------
151 # Constants
152 s, o, a = 'URICUL', 'escherichia coli', 'augmentin'
153
154 # Filter
155 idxs_spec = iti.specimen_code == s
156 idxs_orgs = iti.microorganism_name == o
157 idxs_abxs = iti.antimicrobial_name == a
158 aux = iti[idxs_spec & idxs_orgs & idxs_abxs]
159
160 # --------------
161 # Plot
162 # --------------
163 # Create figure
164 fig, axes = plt.subplots(2, 1, sharex=True,
165 gridspec_kw={'height_ratios': [2, 1]})
166 axes = axes.flatten()
167
168 # Plot line
169 sns.lineplot(x=aux.date_received, y=aux.sari,
170 linewidth=0.75, linestyle='--', #palette="tab10",
171 marker='o', markersize=3, markeredgecolor='k',
172 markeredgewidth=0.5, markerfacecolor=None,
173 alpha=0.5, ax=axes[0])
174
175 # Compute widths
176 widths = [d.days for d in np.diff(aux.date_received.tolist())]
177
178 # Plot bars
179 axes[1].bar(x=aux.date_received, height=aux.freq,
180 width=.8*widths[0], linewidth=0.75, alpha=0.5)
181
182 # Configure
183 axes[0].set(ylim=[-0.1, 1.1],
184 title='[%s, %s, %s] with $%s_{%s}$' % (
185 s, o.upper(), a.upper(), shift, period))
186
187 # Despine
188 sns.despine(bottom=True)
189
190 # Tight layout
191 plt.tight_layout()
192
193 # Show
194 #print("\nTemporal (ITI):")
195 #print(aux)
196 aux
![[URICUL, ESCHERICHIA COLI, AUGMENTIN] with $30D_{30D}$](../../../_images/sphx_glr_plot_step_02_001.png)
Computing ASAI timeseries
Warning
It is important to take into account that computing this index, specially over a period of time, requires a lot of consistent data. Ideally, all the species for the genus of interest should appear on all the time periods.
Once we have computed SARI on a temporal fashion, it is possible
to use such information to compute ASAI in a temporal fashion too.
However, as explained in the previous tutorial, in order to compute
ASAI, we need to at least have columns with the following
information:
antimicrobial
microorganism genus
microorganism species
resistance
Moreover, in this example we will compute the ASAI for each gram_stain category
independently so we will need the microorganism gram stain information too. This
information is available in the registries: pyamr.datasets.registries.
Lets include all this information using the MicroorganismRegistry.
227 # ------------------------------
228 # Include gram stain
229 # ------------------------------
230 # Libraries
231 from pyamr.datasets.registries import MicroorganismRegistry
232
233 # Load registry
234 mreg = MicroorganismRegistry()
235
236 # Format sari dataframe
237 dataframe = iti.copy(deep=True)
238 dataframe = dataframe.reset_index()
239
240 # Create genus and species
241 dataframe[['genus', 'species']] = \
242 dataframe.microorganism_name \
243 .str.capitalize() \
244 .str.split(expand=True, n=1)
245
246 # Combine with registry information
247 dataframe = mreg.combine(dataframe, on='microorganism_name')
248
249 # Fill missing gram stain
250 dataframe.gram_stain = dataframe.gram_stain.fillna('u')
251
252 # Show
253 dataframe.head(4).T
Now that we have the genus, species and gram_stain information,
lets see how to compute ASAI in a temporal fashion with an example. It is
important to highlight that now the date (date_received) is also included
in the groupby parameter when calling the compute method.
For more information see: pyamr.core.asai.ASAI
For more examples see:
269 # -------------------------------------------
270 # Compute ASAI
271 # -------------------------------------------
272 # Import specific libraries
273 from pyamr.core.asai import ASAI
274
275 # Create asai instance
276 asai = ASAI(column_genus='genus',
277 column_specie='species',
278 column_resistance='sari',
279 column_frequency='freq')
280
281 # Compute
282 scores = asai.compute(dataframe,
283 groupby=['date_received',
284 'specimen_code',
285 'antimicrobial_name',
286 'gram_stain'],
287 weights='uniform',
288 threshold=0.5,
289 min_freq=0)
290
291 # Stack
292 scores = scores
293
294 # Show
295 print("\nASAI (overall):")
296 print(scores.unstack())
297 scores.unstack()
c:\users\kelda\desktop\repositories\github\pyamr\main\pyamr\core\asai.py:572: UserWarning:
Extreme resistances [0, 1] were found in the DataFrame. These
rows should be reviewed since these resistances might correspond
to pairs with low number of records.
c:\users\kelda\desktop\repositories\github\pyamr\main\pyamr\core\asai.py:583: UserWarning:
There are NULL values in columns that are required. These
rows will be ignored to safely compute ASAI. Please review
the DataFrame and address this inconsistencies. See below
for more information:
date_received 0
specimen_code 0
antimicrobial_name 0
gram_stain 0
GENUS 0
SPECIE 89
RESISTANCE 0
ASAI (overall):
N_GENUS N_SPECIE ASAI_SCORE
gram_stain n p n p n p
date_received specimen_code antimicrobial_name
2009-01-03 URICUL augmentin 2.0 2.0 2.0 3.0 0.500000 1.00
2009-02-02 URICUL augmentin 2.0 2.0 2.0 3.0 0.500000 0.75
2009-03-04 URICUL augmentin 1.0 2.0 1.0 5.0 1.000000 1.00
2009-04-03 URICUL augmentin 3.0 2.0 3.0 6.0 0.333333 1.00
2009-05-03 URICUL augmentin 3.0 2.0 3.0 5.0 0.666667 1.00
2009-06-02 URICUL augmentin 3.0 2.0 3.0 4.0 0.333333 1.00
2009-07-02 URICUL augmentin 4.0 2.0 4.0 6.0 0.250000 1.00
2009-08-01 URICUL augmentin 1.0 2.0 1.0 5.0 1.000000 1.00
2009-08-31 URICUL augmentin 2.0 2.0 2.0 4.0 0.500000 1.00
2009-09-30 URICUL augmentin 1.0 2.0 1.0 6.0 1.000000 1.00
2009-10-30 URICUL augmentin 3.0 2.0 3.0 5.0 0.333333 0.75
2009-11-29 URICUL augmentin 2.0 2.0 2.0 4.0 0.500000 1.00
2009-12-29 URICUL augmentin 1.0 2.0 1.0 4.0 1.000000 1.00
Let’s plot the evolution for both stains.
304 # Libraries
305 import calendar
306
307 # Month numbers to abbr
308 def month_abbr(v):
309 return [calendar.month_abbr[x] for x in v]
310
311 # --------------
312 # Filter
313 # --------------
314 #
315 s, a = 'URICUL', 'augmentin'
316 # Filter and drop index.
317 scores = scores.filter(like=s, axis=0)
318 scores = scores.filter(like=a, axis=0)
319 scores.index = scores.index.droplevel(level=[1,2])
320
321 # ----------
322 # Plot
323 # ----------
324 # Initialize the matplotlib figure
325 f, ax = plt.subplots(1, figsize=(10, 5))
326
327 # Show
328 sns.lineplot(data=scores, x='date_received', y='ASAI_SCORE',
329 hue='gram_stain', palette="tab10", linewidth=0.75,
330 linestyle='--', marker='o', markersize=3,
331 markeredgecolor='k', markeredgewidth=0.5,
332 markerfacecolor=None, alpha=0.5, ax=ax)#, ax=axes[0])
333
334 # Create aux table for visualization
335 aux = scores[['N_GENUS', 'N_SPECIE']] \
336 .unstack().T.round(0) \
337 .astype(str).replace({'nan': '-'})
338
339 # Rename columns
340 #aux.columns = month_abbr(range(1, len(aux.columns)+1))
341
342 # Draw table
343 table = ax.table(cellText=aux.to_numpy(),
344 rowLabels=aux.index,
345 colLabels=aux.columns.date,
346 cellLoc='center',
347 loc='bottom')
348 table.auto_set_font_size(False)
349 table.set_fontsize(7.5)
350 table.scale(1, 1.2)
351
352 # Sns config
353 sns.despine(left=True, bottom=True)
354
355 # Add a legend and informative axis label
356 ax.set(xlabel='', ylabel='ASAI', xticks=[],
357 title="[%s, %s] with $%s_{%s}$" % (
358 s, a.upper(), shift, period))
359
360
361 # Tight layout()
362 plt.tight_layout()
363
364 # Show
365 plt.show()
366
367 # Show
368 #print("\nASAI (overall):")
369 #print(scores.unstack())
370
371 scores.unstack()
![[URICUL, AUGMENTIN] with $30D_{30D}$](../../../_images/sphx_glr_plot_step_02_002.png)
Further considerations
Warning
Pending!
Total running time of the script: ( 0 minutes 1.421 seconds)