Note
Click here to download the full example code
05c. Custom using swarmplot
This script demonstrates how to build a custom SHAP visualization
for sequential data using seaborn.swarmplot. This technique
creates a detailed, per-feature view that shows the distribution
of SHAP values at each timestep, offering a high-density
alternative to standard summary plots.
The script’s workflow includes:
Loading & Subsetting Data: It loads a pre-computed, tidy DataFrame of SHAP values and subsets it to ensure the computationally intensive swarmplot runs efficiently.
Per-Feature Visualization: The script iterates through each feature, generating a separate swarmplot to clearly display its specific impact across the entire time sequence.
Custom Value-Based Coloring: It implements a custom coloring function to tint each point based on its original feature value, adding a color bar to provide the rich context found in native SHAP plots.
Plot Customization: It showcases how to fine-tune the plot’s appearance, including normalizing the y-axis and managing legends for a polished final output.
This example is perfect for users who need to visualize the precise distribution of feature impacts over time, though it is best suited for smaller datasets where the swarmplot can arrange points without significant overlap.
33 # Libraries
34 import shap
35 import numpy as np
36 import pandas as pd
37 import seaborn as sns
38
39 import matplotlib.pyplot as plt
40 import matplotlib as mpl
41 import matplotlib.colorbar
42 import matplotlib.colors
43 import matplotlib.cm
44
45 from mpl_toolkits.axes_grid1 import make_axes_locatable
46
47 try:
48 __file__
49 TERMINAL = True
50 except:
51 TERMINAL = False
52
53
54 # ------------------------
55 # Methods
56 # ------------------------
57 def scalar_colormap(values, cmap, vmin, vmax):
58 """This method creates a colormap based on values.
59
60 Parameters
61 ----------
62 values : array-like
63 The values to create the corresponding colors
64
65 cmap : str
66 The colormap
67
68 vmin, vmax : float
69 The minimum and maximum possible values
70
71 Returns
72 -------
73 scalar colormap
74 """
75 # Create scalar mappable
76 norm = mpl.colors.Normalize(vmin=vmin, vmax=vmax, clip=True)
77 mapper = mpl.cm.ScalarMappable(norm=norm, cmap=cmap)
78 # Get color map
79 colormap = sns.color_palette([mapper.to_rgba(i) for i in values])
80 # Return
81 return colormap, norm
82
83
84 def scalar_palette(values, cmap, vmin, vmax):
85 """This method creates a colorpalette based on values.
86
87 Parameters
88 ----------
89 values : array-like
90 The values to create the corresponding colors
91
92 cmap : str
93 The colormap
94
95 vmin, vmax : float
96 The minimum and maximum possible values
97
98 Returns
99 -------
100 scalar colormap
101
102 """
103 # Create a matplotlib colormap from name
104 # cmap = sns.light_palette(cmap, reverse=False, as_cmap=True)
105 cmap = sns.color_palette(cmap, as_cmap=True)
106 # Normalize to the range of possible values from df["c"]
107 norm = mpl.colors.Normalize(vmin=vmin, vmax=vmax)
108 # Create a color dictionary (value in c : color from colormap)
109 colors = {}
110 for cval in values:
111 colors.update({cval: cmap(norm(cval))})
112 # Return
113 return colors, norm
114
115 def load_shap_file():
116 """Load shap file.
117
118 .. note: The timestep does not indicate time step but matrix
119 index index. Since the matrix index for time steps
120 started in negative t=-T and ended in t=0 the
121 transformation should be taken into account.
122
123 """
124 from pathlib import Path
125 # Load data
126 path = Path('../../datasets/shap/')
127 data = pd.read_csv(path / 'shap.csv')
128 data = data.iloc[:, 1:]
129 data = data.rename(columns={'timestep': 'indice'})
130 data['timestep'] = data.indice - (data.indice.nunique() - 1)
131 return data
132
133
134
135 # ------------------------------------------------------
136 # Main
137 # ------------------------------------------------------
138 # Configuration
139 cmap_name = 'coolwarm' # colormap name
140 norm_shap = True
141
142 # Load data
143 data = load_shap_file()
144
145 # Filter so that it is less computationally expensive
146 data = data[data['sample'] < 100]
147
148 # Show
149 if TERMINAL:
150 print("\nShow:")
151 print(data)
Let’s see how data looks like
155 data.head(10)
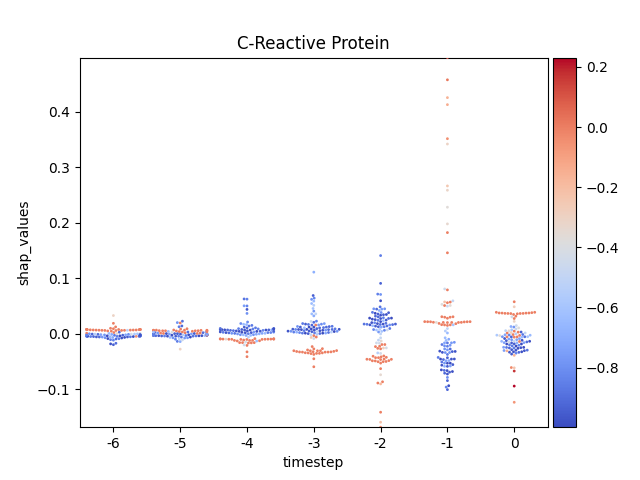
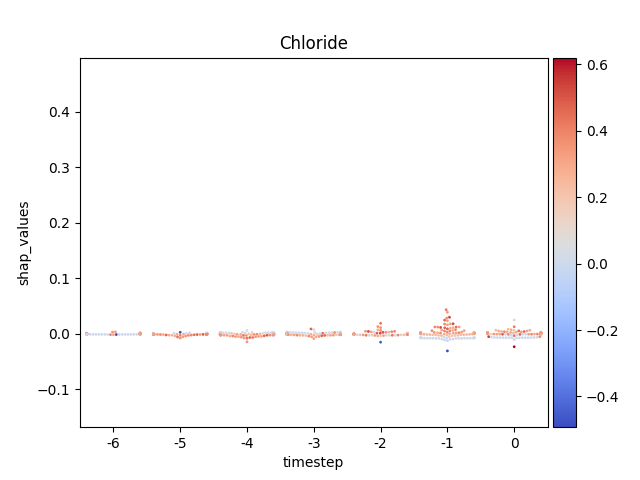
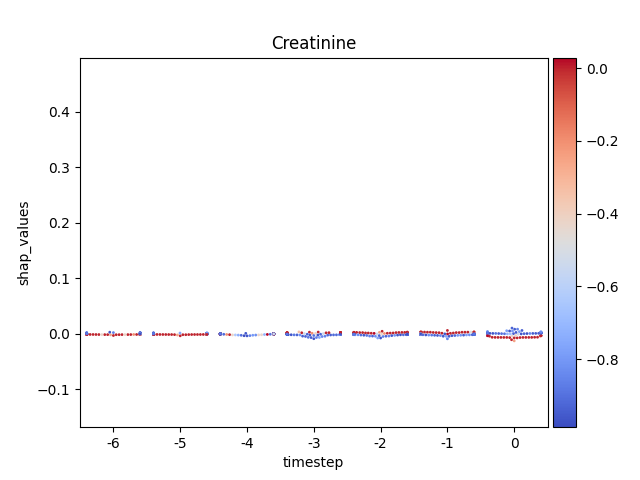
Display using sns.swarmplot
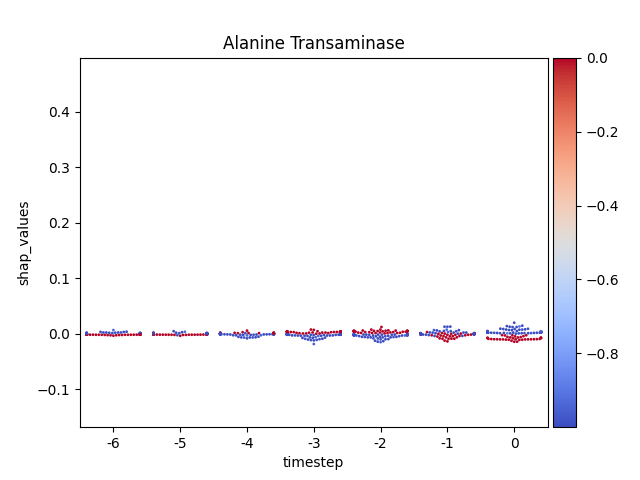
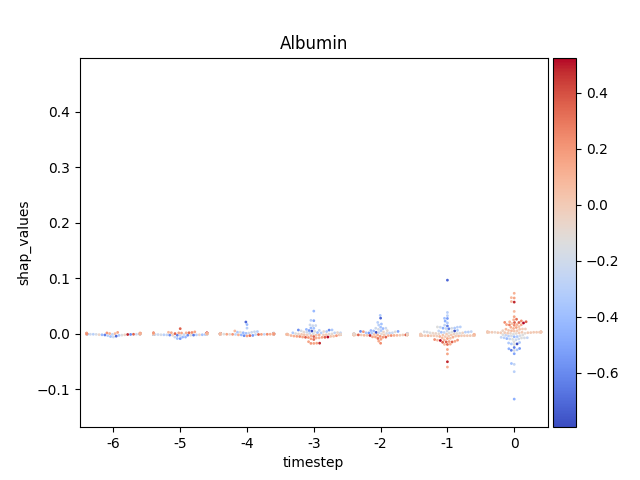
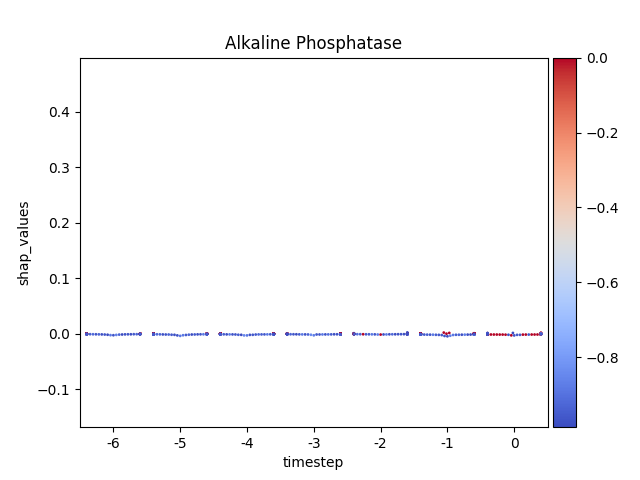
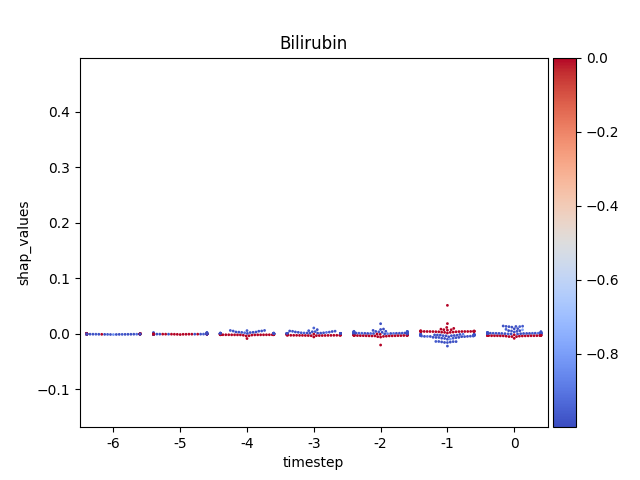
Warning
This method seems to be quite slow.
Note
y-axis has been ‘normalized’
166 def add_colorbar(fig, cmap, norm):
167 """"""
168 divider = make_axes_locatable(plt.gca())
169 ax_cb = divider.new_horizontal(size="5%", pad=0.05)
170 fig.add_axes(ax_cb)
171 cb1 = matplotlib.colorbar.ColorbarBase(ax_cb,
172 cmap=cmap, norm=norm, orientation='vertical')
173
174
175 # Loop
176 for i, (name, df) in enumerate(data.groupby('features')):
177
178 # Get colormap
179 values = df.feature_values
180 cmap, norm = scalar_palette(values=values,
181 cmap=cmap_name, vmin=values.min(),
182 vmax=values.max())
183
184 # Display
185 fig, ax = plt.subplots()
186 ax = sns.swarmplot(x='timestep',
187 y='shap_values',
188 hue='feature_values',
189 palette=cmap,
190 data=df,
191 size=2,
192 ax=ax)
193
194 # Format figure
195 plt.title(name)
196 plt.legend([], [], frameon=False)
197
198 if norm_shap:
199 plt.ylim(data.shap_values.min(),
200 data.shap_values.max())
201
202 # Invert x axis (if no negative timesteps)
203 #ax.invert_xaxis()
204
205 # Create colormap (fix for old versions of mpl)
206 cmap = matplotlib.cm.get_cmap(cmap_name)
207
208 # Add colorbar
209 add_colorbar(plt.gcf(), cmap, norm)
210
211 # Show only first N
212 if int(i) > 5:
213 break
214
215 # Show
216 plt.show()
Out:
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
59.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
37.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
53.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
21.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
19.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
53.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
49.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
21.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
8.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
30.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
34.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
8.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
32.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
18.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
8.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
17.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\virtualenvs\venv-py311-psc\Lib\site-packages\seaborn\categorical.py:3399: UserWarning:
6.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:206: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
C:\Users\kelda\Desktop\repositories\github\python-spare-code\main\examples\shap\plot_main05_swarmplot.py:216: UserWarning:
FigureCanvasAgg is non-interactive, and thus cannot be shown
Total running time of the script: ( 0 minutes 11.309 seconds)