Note
Go to the end to download the full example code
Collateral Sensitivity Index (ACSI)
The Collateral Sensitivity Index - ACSI …
Warning
Pending…!
First, lets create the susceptibility test records dataset
14 # Libraries
15 import pandas as pd
16
17 from pyamr.datasets.load import fixture
18
19 # ----------------------------------
20 # Create data
21 # ----------------------------------
22 # Load the fixture_05.csv file.
23 susceptibility = fixture(name='fixture_06.csv')
24
25 # Format DataFrame
26 susceptibility.SENSITIVITY = \
27 susceptibility.SENSITIVITY.replace({
28 'resistant': 'R',
29 'intermediate': 'I',
30 'sensitive': 'S'
31 })
Lets see the susceptibility test records
35 susceptibility.head(10)
Lets compute the Antimicrobial Collateral Sensitivity Index or ACSI
42 # ------------------------------------------
43 # Compute the index
44 # ------------------------------------------
45 # Libraries
46 from pyamr.core.acsi import ACSI
47
48 # Compute index
49 contingency, combinations = \
50 ACSI().compute(susceptibility,
51 groupby=[
52 'DATE',
53 'SPECIMEN',
54 'MICROORGANISM'
55 ],
56 return_combinations=True)
57
58 # Show
59 print("\nCombinations:")
60 print(combinations)
61 print("\nContingency:")
62 print(contingency)
Combinations:
DATE SPECIMEN MICROORGANISM LAB_NUMBER ANTIMICROBIAL_x ANTIMICROBIAL_y SENSITIVITY_x SENSITIVITY_y class
0 2021-01-01 BLDCUL ECOL lab1 AAUG ACIP S S SS
1 2021-01-01 BLDCUL ECOL lab2 AAUG ACIP S R SR
2 2021-01-01 BLDCUL ECOL lab3 AAUG ACIP S R SR
3 2021-01-01 BLDCUL ECOL lab4 AAUG ACIP R R RR
4 2021-01-02 BLDCUL ECOL lab5 AAUG ACIP S S SS
5 2021-01-02 BLDCUL ECOL lab6 AAUG ACIP S R SR
6 2021-01-02 BLDCUL ECOL lab7 AAUG ACIP R R RR
7 2021-01-03 BLDCUL ECOL lab8 AAUG ACIP S I SI
8 2021-01-03 BLDCUL ECOL lab9 AAUG ACIP R R RR
9 2021-01-03 BLDCUL SAUR lab9 ACIP ACIP R R RR
10 2021-01-04 URICUL ECOL lab10 AAUG ACIP R S RS
11 2021-01-04 URICUL SAUR lab10 AAUG APEN R R RR
Contingency:
class RR RS SI SR SS acsi
DATE SPECIMEN MICROORGANISM ANTIMICROBIAL_x ANTIMICROBIAL_y
2021-01-01 BLDCUL ECOL AAUG ACIP 1.0 NaN NaN 2.0 1.0 0.2027
2021-01-02 BLDCUL ECOL AAUG ACIP 1.0 NaN NaN 1.0 1.0 0.3662
2021-01-03 BLDCUL ECOL AAUG ACIP 1.0 NaN 1.0 NaN NaN 0.0000
SAUR ACIP ACIP 1.0 NaN NaN NaN NaN 0.0000
2021-01-04 URICUL ECOL AAUG ACIP NaN 1.0 NaN NaN NaN 0.0000
SAUR AAUG APEN 1.0 NaN NaN NaN NaN 0.0000
Lets see the contingency matrix
67 # Rename for docs display
68 rename = {
69 'MICROORGANISM': 'ORGANISM',
70 'ANTIMICROBIAL_x': 'ANTIBIOTIC_x',
71 'ANTIMICROBIAL_y': 'ANTIBIOTIC_y'
72 }
73
74 # Show
75 contingency \
76 .reset_index() \
77 .rename(columns=rename) \
78 .round(decimals=3)
Lets compute and visualise the overall contingency matrix
85 # Lets compute the overall index
86 contingency = ACSI().compute(
87 combinations.reset_index(),
88 groupby=[],
89 flag_combinations=True,
90 return_combinations=False
91 )
92
93 # Show
94 print(contingency)
95
96 # Display
97 import numpy as np
98 import seaborn as sns
99 import matplotlib.pyplot as plt
100
101 # Create index with all pairs
102 index = pd.MultiIndex.from_product(
103 [susceptibility.ANTIMICROBIAL.unique(),
104 susceptibility.ANTIMICROBIAL.unique()]
105 )
106
107 # Reformat
108 aux = contingency['acsi'] \
109 .reindex(index, fill_value=np.nan)\
110 .unstack()
111
112 # Display
113 sns.heatmap(data=aux * 100, annot=True, linewidth=.5,
114 cmap='coolwarm', vmin=-70, vmax=70, center=0,
115 square=True)
116
117 # Show
118 plt.show()
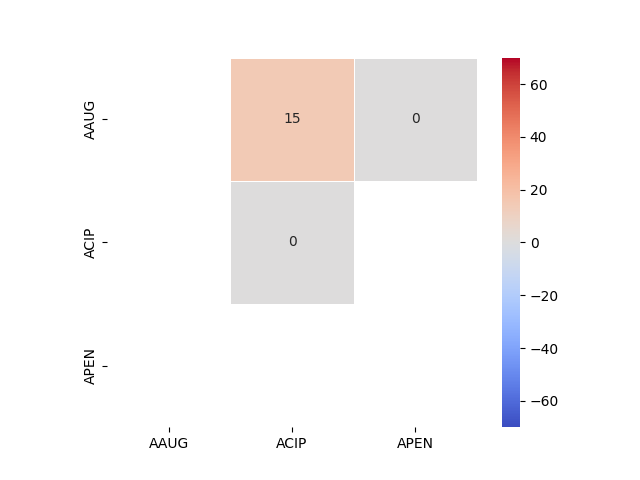
class RR RS SI SR SS acsi
ANTIMICROBIAL_x ANTIMICROBIAL_y
AAUG ACIP 3.0 1.0 1.0 3.0 2.0 0.1469
APEN 1.0 NaN NaN NaN NaN 0.0000
ACIP ACIP 1.0 NaN NaN NaN NaN 0.0000
Lets compute and visualise the temporal contingency matrix
123 # Lets compute the overall index
124 contingency = ACSI().compute(
125 combinations.reset_index(),
126 groupby=['DATE'],
127 flag_combinations=True,
128 return_combinations=False)
129
130 # Show
131 print(contingency)
132
133 # .. note:: Look for a nice visualisation of a matrix over time. In
134 # addition to python libraries such as plotly, animations,
135 # ... we can also check for js libraries such as Apache
136 # echarts.
class RR RS SI SR SS acsi
DATE ANTIMICROBIAL_x ANTIMICROBIAL_y
2021-01-01 AAUG ACIP 1.0 NaN NaN 2.0 1.0 0.2027
2021-01-02 AAUG ACIP 1.0 NaN NaN 1.0 1.0 0.3662
2021-01-03 AAUG ACIP 1.0 NaN 1.0 NaN NaN 0.0000
ACIP ACIP 1.0 NaN NaN NaN NaN 0.0000
2021-01-04 AAUG ACIP NaN 1.0 NaN NaN NaN 0.0000
APEN 1.0 NaN NaN NaN NaN 0.0000
Total running time of the script: ( 0 minutes 0.149 seconds)