Note
Go to the end to download the full example code
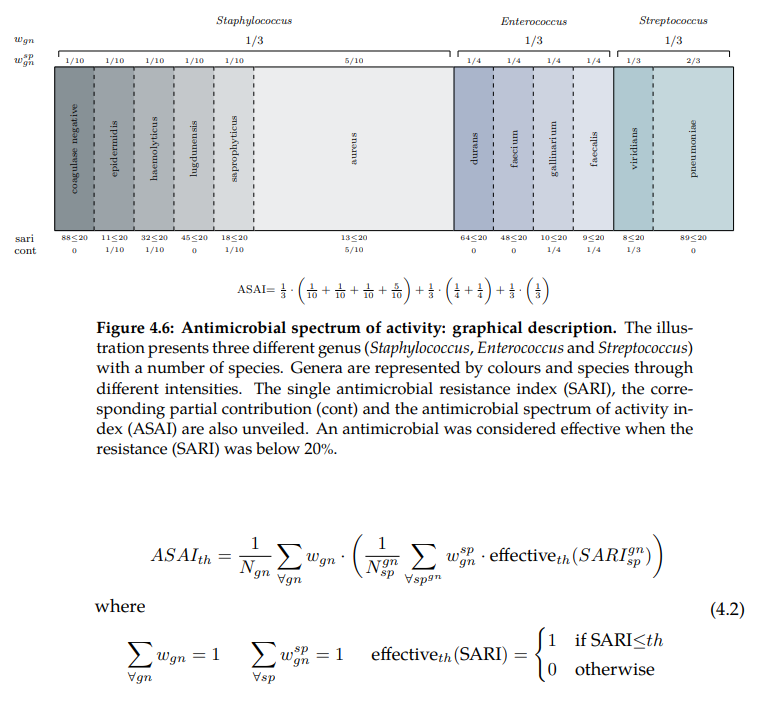
Spectrum of Activity (ASAI)
Warning
There is an error in the Figure displayed below. The contribution (cont) value for Staphylococcus haemolyticus should be 0 instead of 1/10 because the sari index is higher than the threshold (32 > 20) and therefore the antimicrobial is not considered effective.
The Antimicrobial Spectrum of Activity Index or ASAI refers to the range of microbe species that are
susceptible to these agents and therefore can be treated. In general, antimicrobial agents
are classified into broad, intermediate or narrow spectrum. Broad spectrum antimicrobials
are active against both Gram-positive and Gram-negative bacteria. In contrast, narrow
spectrum antimicrobials have limited activity and are effective only against particular
species of bacteria. While these profiles appeared in the mid-1950s, little effort has been
made to define them. Furthermore, such ambiguous labels are overused for different
and even contradictory purposes.
For more information see: pyamr.core.asai.ASAI
28 # Import libraries
29 import sys
30 import pandas as pd
31 import numpy as np
32 import seaborn as sns
33 import matplotlib as mpl
34 import matplotlib.pyplot as plt
35
36 # Import specific libraries
37 from pyamr.core.asai import ASAI
38 from pyamr.datasets.load import fixture
39 from pyamr.utils.plot import scalar_colormap
40
41 # Configure seaborn style (context=talk)
42 sns.set(style="white")
43
44 # Set matplotlib
45 mpl.rcParams['xtick.labelsize'] = 9
46 mpl.rcParams['ytick.labelsize'] = 9
47 mpl.rcParams['axes.titlesize'] = 11
48 mpl.rcParams['legend.fontsize'] = 9
49
50 # Pandas configuration
51 pd.set_option('display.max_colwidth', 40)
52 pd.set_option('display.width', 300)
53 pd.set_option('display.precision', 4)
54
55 # Numpy configuration
56 np.set_printoptions(precision=2)
57
58
59 # ---------------------
60 # Create data
61 # ---------------------
62 # Load data
63 dataframe = fixture(name='fixture_04.csv')
64
65 # Show
66 print("Summary:")
67 print(dataframe)
Summary:
GENUS SPECIE ANTIBIOTIC GRAM RESISTANCE FREQUENCY THRESHOLD W_SPECIE W_GENUS
0 Staphylococcus coagulase negative ANTIBIOTIC_1 P 0.88 1 0.2 0.1000 0.3333
1 Staphylococcus epidermidis ANTIBIOTIC_1 P 0.11 1 0.2 0.1000 0.3333
2 Staphylococcus haemolyticus ANTIBIOTIC_1 P 0.32 1 0.2 0.1000 0.3333
3 Staphylococcus lugdumensis ANTIBIOTIC_1 P 0.45 1 0.2 0.1000 0.3333
4 Staphylococcus saporphyticus ANTIBIOTIC_1 P 0.18 1 0.2 0.1000 0.3333
5 Staphylococcus aureus ANTIBIOTIC_1 P 0.13 5 0.2 0.5000 0.3333
6 Enterococcus durans ANTIBIOTIC_1 N 0.64 1 0.2 0.2500 0.3333
7 Enterococcus faecium ANTIBIOTIC_1 N 0.48 1 0.2 0.2500 0.3333
8 Enterococcus gallinarium ANTIBIOTIC_1 N 0.10 1 0.2 0.2500 0.3333
9 Enterococcus faecalis ANTIBIOTIC_1 N 0.09 1 0.2 0.2500 0.3333
10 Streptococcus viridians ANTIBIOTIC_1 P 0.08 1 0.2 0.3333 0.3333
11 Streptococcus pneumoniae ANTIBIOTIC_1 P 0.89 2 0.2 0.6667 0.3333
12 Staphylococcus coagulase negative ANTIBIOTIC_2 P 0.88 1 0.2 0.1000 0.3333
13 Staphylococcus epidermidis ANTIBIOTIC_2 P 0.11 1 0.5 0.1000 0.3333
14 Staphylococcus haemolyticus ANTIBIOTIC_2 P 0.32 1 0.5 0.1000 0.3333
15 Staphylococcus lugdumensis ANTIBIOTIC_2 P 0.45 1 0.5 0.1000 0.3333
16 Staphylococcus saporphyticus ANTIBIOTIC_2 P 0.18 1 0.2 0.1000 0.3333
17 Staphylococcus aureus ANTIBIOTIC_2 P 0.13 5 0.2 0.5000 0.3333
18 Enterococcus durans ANTIBIOTIC_2 N 0.64 1 0.2 0.2500 0.3333
19 Enterococcus faecium ANTIBIOTIC_2 N 0.48 1 0.2 0.2500 0.3333
20 Enterococcus gallinarium ANTIBIOTIC_2 N 0.10 1 0.5 0.2500 0.3333
21 Enterococcus faecalis ANTIBIOTIC_2 N 0.09 1 0.5 0.2500 0.3333
22 Streptococcus viridians ANTIBIOTIC_2 P 0.08 1 0.5 0.3333 0.3333
23 Streptococcus pneumoniae ANTIBIOTIC_2 P 0.89 2 0.5 0.6667 0.3333
Lets see the dataframe
71 dataframe
Lets use the ASAI object
78 # -------------------------------
79 # Create antimicrobial spectrum
80 # -------------------------------
81 # Create antimicrobial spectrum of activity instance
82 asai = ASAI(column_genus='GENUS',
83 column_specie='SPECIE',
84 column_resistance='RESISTANCE',
85 column_frequency='FREQUENCY',
86 column_threshold='THRESHOLD',
87 column_wgenus='W_GENUS',
88 column_wspecie='W_SPECIE')
89
90 # Compute ASAI with uniform weights
91 scores1 = asai.compute(dataframe,
92 groupby=['ANTIBIOTIC', 'GRAM'],
93 weights='uniform',
94 threshold=None,
95 min_freq=0).unstack()
96
97 # compute ASAI with frequency weights
98 scores2 = asai.compute(dataframe,
99 groupby=['ANTIBIOTIC', 'GRAM'],
100 weights='frequency',
101 threshold=None,
102 min_freq=0).unstack()
Lets see the ASAI with uniform weights
106 scores1
Lets see the ASAI with frequency weights
110 scores2
Lets display the information graphically
116 def plot_graph_asai(scores):
117 """Display ASAI as a horizontal bars
118 """
119 # Variables to plot.
120 x = scores.index.values
121 y_n = scores['ASAI_SCORE']['N'].values
122 y_p = scores['ASAI_SCORE']['P'].values
123
124 # Constants
125 colormap_p = scalar_colormap(y_p, cmap='Blues', vmin=-0.1, vmax=1.1)
126 colormap_n = scalar_colormap(y_n, cmap='Reds', vmin=-0.1, vmax=1.1)
127
128 # Create figure
129 f, ax = plt.subplots(1, 1, figsize=(8, 0.5))
130
131 # Plot
132 sns.barplot(x=y_p, y=x, palette=colormap_p, ax=ax, orient='h',
133 saturation=0.5, label='Gram-positive')
134 sns.barplot(x=-y_n, y=x, palette=colormap_n, ax=ax, orient='h',
135 saturation=0.5, label='Gram-negative')
136
137 # Configure
138 sns.despine(bottom=True)
139
140 # Configure
141 ax.set_xlim([-1, 1])
142 ax.legend(loc='center left')
143
144 # -----------------------------
145 # Plot
146 # -----------------------------
147 # Display bar graphs
148 plot_graph_asai(scores1)
149 plot_graph_asai(scores2)
150
151 # Display
152 plt.show()
Total running time of the script: ( 0 minutes 0.197 seconds)

