Note
Click here to download the full example code
Collateral Sensitivity Index
In order to run the script in such a way that we can profile the time used for each method, statement, .. use the following command:
$ python -m cProfile -s cumtime plot_collateral_sensitivity.py > outcome.csv
11 # Libraries
12 import numpy as np
13 import pandas as pd
14 import seaborn as sns
15 import matplotlib as mpl
16 import matplotlib.pyplot as plt
17
18 #
19 from pathlib import Path
20 from datetime import datetime
21 from itertools import combinations
22 from mic import mutual_info_matrix_v3
23
24 # See https://matplotlib.org/devdocs/users/explain/customizing.html
25 mpl.rcParams['axes.titlesize'] = 8
26 mpl.rcParams['axes.labelsize'] = 8
27 mpl.rcParams['xtick.labelsize'] = 8
28 mpl.rcParams['ytick.labelsize'] = 8
29
30 def collateral_resistance_index(m):
31 """Collateral Resistance Index
32
33 The collateral resistance index is based on the mutual
34 information matrix. This implementation assumes there
35 are two classes resistant (R) and sensitive (S).
36
37 Parameters
38 ----------
39 m: np.array
40 A numpy array with the mutual information matrix.
41
42 Returns
43 -------
44 """
45 return (m[0, 0] + m[1, 1]) - (m[0, 1] + m[1, 0])
46
47 def CRI(x, func):
48 ct = np.array([[x.SS, x.SR], [x.RS, x.RR]])
49 m = func(ct=ct)
50 return collateral_resistance_index(m)
51
52
53
54 def combo_v1():
55 # Build combination
56 c = pd.DataFrame()
57 for i, g in df.groupby(['o', 's']):
58 for index in list(combinations(g.index, 2)):
59 i, j = index
60 s = pd.Series({
61 'o': df.loc[i, 'o'],
62 's': df.loc[i, 's'],
63 'ax': df.loc[i, 'a'],
64 'ay': df.loc[j, 'a'],
65 'rx': df.loc[i, 'r'],
66 'ry': df.loc[j, 'r']
67 })
68 c = pd.concat([c, s.to_frame().T])
69 # c.append(s)
70
71 """
72 # Build combination
73 c = pd.DataFrame()
74 for i, g in data.groupby(['specimen_code',
75 'microorganism_code',
76 'laboratory_number']):
77 for index in list(combinations(g.index, 2)):
78 i, j = index
79 s = pd.Series({
80 'o': data.loc[i, 'microorganism_code'],
81 's': data.loc[i, 'laboratory_number'],
82 'ax': data.loc[i, 'antimicrobial_code'],
83 'ay': data.loc[j, 'antimicrobial_code'],
84 'rx': data.loc[i, 'sensitivity'],
85 'ry': data.loc[j, 'sensitivity']
86 })
87 c = pd.concat([c, s.to_frame().T])
88
89
90 # Add class
91 c['class'] = c.rx + c.ry
92 """
93 return c
94
95
96 def create_df_combo_v1(d, col_o='o', # organism
97 col_s='s', # sample
98 col_a='a', # antimicrobial
99 col_r='r'): # outcome / result
100 """
101
102 .. note:: There might be an issue if there are two different outcomes
103 for the same record. For example, a susceptibility test
104 record for penicillin (APEN) with R and another one with
105 S. Warn of this issue if it appears!
106
107 :param d:
108 :param col_o:
109 :param col_s:
110 :param col_a:
111 :param col_r:
112 :return:
113 """
114
115 # This is innefficient!
116
117 # Build combination
118 c = []
119 for i, g in d.groupby([col_s, col_o]):
120 for x, y in combinations(g.sort_values(by=col_a).index, 2):
121 s = pd.Series({
122 'o': g.loc[x, col_o],
123 's': g.loc[x, col_s],
124 'ax': g.loc[x, col_a],
125 'ay': g.loc[y, col_a],
126 'rx': g.loc[x, col_r],
127 'ry': g.loc[y, col_r]
128 })
129 c.append(s)
130
131
132 # Concatenate
133 c = pd.concat(c, axis=1).T
134
135 # Add class
136 c['class'] = c.rx + c.ry
137
138 # Return
139 return c
140
141
142 def create_combinations_v1(d, col_specimen='s',
143 col_lab_id='l',
144 col_microorganism='o',
145 col_antimicrobial='a',
146 col_result='r'):
147 """Creates the dataframe with all combinations.
148
149 Parameters
150 ----------
151
152 Returns
153 --------
154 """
155 # Initialize
156 c = []
157
158 # Loop
159 for i, g in d.groupby([col_specimen,
160 col_microorganism,
161 col_lab_id]):
162 for x, y in combinations(g.sort_values(by=col_antimicrobial).index, 2):
163 c.append({
164 'specimen': g.loc[x, col_specimen],
165 'lab_id': g.loc[x, col_lab_id],
166 'o': g.loc[x, col_microorganism],
167 'ax': g.loc[x, col_antimicrobial],
168 'ay': g.loc[y, col_antimicrobial],
169 'rx': g.loc[x, col_result],
170 'ry': g.loc[y, col_result]
171 })
172
173 # Create DataFrame
174 c = pd.DataFrame(c)
175 # Add class
176 c['class'] = c.rx + c.ry
177 # Return
178 return c
179
180
181 def create_combinations_v2(d, col_o='o',
182 col_s='s', col_a='a', col_r='r'):
183 """Creates the dataframe with all combinations.
184
185 .. note:: There might be an issue if there are two different outcomes
186 for the same record. For example, a susceptibility test
187 record for penicillin (APEN) with R and another one with
188 S. Warn of this issue if it appears!
189
190 Parameters
191 ----------
192
193 Returns
194 --------
195
196 """
197 # Initialize
198 c = pd.DataFrame()
199
200 # Loop
201 for i, g in d.groupby([col_s, col_o]):
202
203 aux = []
204 for x, y in combinations(g.sort_values(by=col_a).index, 2):
205 aux.append({
206 'ax': g.loc[x, col_a],
207 'ay': g.loc[y, col_a],
208 'rx': g.loc[x, col_r],
209 'ry': g.loc[y, col_r]
210 })
211 aux = pd.DataFrame(aux)
212 aux['s'] = i[0]
213 aux['o'] = i[1]
214
215 # Concatenate
216 c = pd.concat([c, aux], axis=0)
217
218 # Add class
219 c['class'] = c.rx + c.ry
220
221 # Return
222 return c
A basic example
Note that the columns names are the initial for the following full names: s=specimen, l=laboratory sample, o=organism, a=antimicrobial and r=result
231 # Create matrix
232 data = [
233 ['s1', 'l1', 'o1', 'a1', 'S'],
234 ['s1', 'l1', 'o1', 'a2', 'S'],
235 ['s1', 'l1', 'o1', 'a3', 'R'],
236 ['s1', 'l2', 'o1', 'a1', 'S'],
237 ['s1', 'l2', 'o1', 'a2', 'S'],
238 ['s1', 'l2', 'o1', 'a3', 'R'],
239 ['s1', 'l2', 'o1', 'a4', 'R'],
240 ['s1', 'l3', 'o1', 'a1', 'R'],
241 ['s1', 'l3', 'o1', 'a2', 'S'],
242 ['s1', 'l4', 'o1', 'a2', 'R'],
243 ['s1', 'l4', 'o1', 'a1', 'S'],
244 ['s1', 'l5', 'o1', 'a5', 'S'],
245 ['s1', 'l6', 'o1', 'a4', 'S'],
246 ['s1', 'l5', 'o1', 'a2', 'S'],
247 ]
248
249 # Create DataFrame
250 df = pd.DataFrame(data,
251 columns=['s', 'l', 'o', 'a', 'r'])
252
253 # Show
254 print("\nData:")
255 print(df)
256
257 # Create combo
258 c = create_combinations_v1(df)
259
260 # Show
261 print("\nCombinations (within isolates):")
262 print(c)
263
264 # Build contingency
265 r = c.groupby(['ax', 'ay', 'class']).size().unstack()
266
267 # Show
268 print("\nContingency:")
269 print(r)
270
271 # Compute CRI
272 r['MIS'] = r.apply(CRI, args=(mutual_info_matrix_v3,), axis=1)
273
274 # Show
275 print("\n" + "="*80 + "\nExample 1\n" + "="*80)
276 print("\nResult")
277 print(r)
278
279 # Create index with all pairs
280 index = pd.MultiIndex.from_product(
281 [df.a.unique(), df.a.unique()]
282 )
283
284 # Reformat
285 aux = r['MIS'] \
286 .reindex(index, fill_value=np.nan)\
287 .unstack()
288
289 # Display
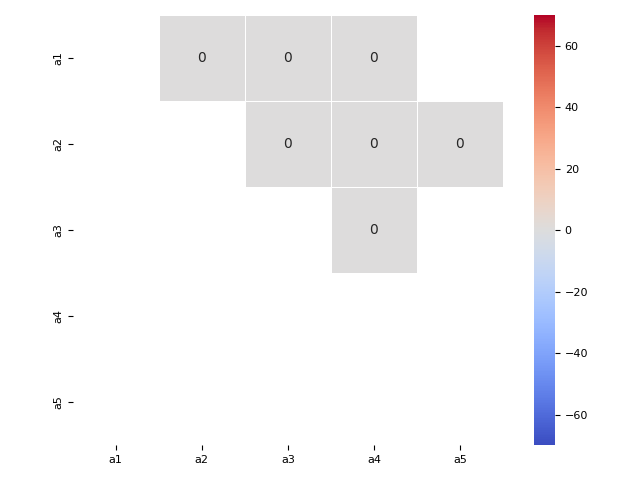
290 sns.heatmap(data=aux*100, annot=True, linewidth=.5,
291 cmap='coolwarm', vmin=-70, vmax=70, center=0,
292 square=True)
293
294 # Show
295 plt.tight_layout()
296 #plt.show()
297
298
299
300
301 def load_susceptibility_nhs(**kwargs):
302 """Load and format MIMIC microbiology data.
303
304 Parameters
305 ----------
306 **kwargs: dict-like
307 The arguments as used in pandas read_csv function
308
309 Returns
310 --------
311 """
312 # Load data
313 path = Path('../../datasets/susceptibility-nhs/')
314 path = path / 'susceptibility-v0.0.1'
315
316 data = pd.concat([
317 pd.read_csv(f, **kwargs)
318 for f in Path(path).glob('susceptibility-*.csv')])
319
320 # Format data
321 data.sensitivity = data.sensitivity \
322 .replace({
323 'sensitive': 'S',
324 'resistant': 'R',
325 'intermediate': 'I',
326 'highly resistant': 'HR'
327 })
328
329 # Select specimen
330 # data = data[data.specimen_code.isin(['URICUL'])]
331 # data = data[data.microorganism_code.isin(['SAUR', 'ECOL', 'PAER'])]
332 # data = data[data.sensitivity.isin(['R', 'S'])]
333 # data = data[data.laboratory_number.isin(['H1954180', 'M1596362'])]
334
335 data = data[data.sensitivity.isin(['R', 'S', 'I', 'HR'])]
336
337 # .. note:: For some reason, for the same specimen and antimicrobial
338 # there are sometimes contradictory outcomes (e.g. R and S)
339 # so we are removing this by keeping the last.
340
341 # Keep only last/first specimen (sometimes repeated)
342 subset = data.columns.tolist()
343 subset = subset.remove('sensitivity')
344 data = data.drop_duplicates(subset=subset, keep='last')
345
346 # Further cleaning
347
348 # Return
349 return data
350
351
352 def load_susceptibility_mimic(**kwargs):
353 """Load and format MIMIC microbiology data.
354
355 Parameters
356 ----------
357 **kwargs: dict-like
358 The arguments as used in pandas read_csv function
359
360 Returns
361 --------
362 """
363 # Load data
364 path = Path('../../datasets/susceptibility-mimic/')
365 path = path / 'microbiologyevents.csv'
366 data = pd.read_csv(path, **kwargs)
367
368 # Format data
369 data = data.rename(columns={
370 'micro_specimen_id': 'laboratory_number',
371 'spec_type_desc': 'specimen_code',
372 'org_name': 'microorganism_code',
373 'ab_name': 'antimicrobial_code',
374 'interpretation': 'sensitivity'
375 })
376
377 # Keep only last/first specimen
378
379 # Remove inconsistent records, for example if for an specimen there are two
380 # rows for the same antimicrobial. Or even worse, these two rows are
381 # contradictory (e.g. R and S)
382
383 # Other cleaning.
384
385 # Return
386 return data
387
388
389 """
390 # Load data
391 #data = load_susceptibility_mimic()
392 data = load_susceptibility_nhs()
393
394 # Create combo
395 c = create_combinations_v1(data,
396 col_specimen='specimen_code',
397 col_lab_id='laboratory_number',
398 col_microorganism='microorganism_code',
399 col_antimicrobial='antimicrobial_code',
400 col_result='sensitivity')
401
402 # Create folder if it does not exist.
403 today = datetime.now().strftime("%Y%m%d-%H%M%S")
404 path = Path('./outputs/cri/') / today
405 Path(path).mkdir(parents=True, exist_ok=True)
406
407 # Save combinations file.
408 c.to_csv(path / 'combinations.csv')
409
410 # Build contingency
411 r = c.groupby(['specimen', 'o', 'ax', 'ay', 'class']).size().unstack()
412
413 # Compute CRI
414 r['MIS'] = r.fillna(0) \
415 .apply(CRI, args=(mutual_info_matrix_v3,), axis=1)
416
417 # Show
418 print("\n" + "="*80 + "\nExample 2\n" + "="*80)
419 print("\nResult")
420 print(r)
421
422 # Save collateral sensitivity index file.
423 r.to_csv(path / 'contingency.csv')
424 """
Out:
Data:
s l o a r
0 s1 l1 o1 a1 S
1 s1 l1 o1 a2 S
2 s1 l1 o1 a3 R
3 s1 l2 o1 a1 S
4 s1 l2 o1 a2 S
5 s1 l2 o1 a3 R
6 s1 l2 o1 a4 R
7 s1 l3 o1 a1 R
8 s1 l3 o1 a2 S
9 s1 l4 o1 a2 R
10 s1 l4 o1 a1 S
11 s1 l5 o1 a5 S
12 s1 l6 o1 a4 S
13 s1 l5 o1 a2 S
Combinations (within isolates):
specimen lab_id o ax ay rx ry class
0 s1 l1 o1 a1 a2 S S SS
1 s1 l1 o1 a1 a3 S R SR
2 s1 l1 o1 a2 a3 S R SR
3 s1 l2 o1 a1 a2 S S SS
4 s1 l2 o1 a1 a3 S R SR
5 s1 l2 o1 a1 a4 S R SR
6 s1 l2 o1 a2 a3 S R SR
7 s1 l2 o1 a2 a4 S R SR
8 s1 l2 o1 a3 a4 R R RR
9 s1 l3 o1 a1 a2 R S RS
10 s1 l4 o1 a1 a2 S R SR
11 s1 l5 o1 a2 a5 S S SS
Contingency:
class RR RS SR SS
ax ay
a1 a2 NaN 1.0 1.0 2.0
a3 NaN NaN 2.0 NaN
a4 NaN NaN 1.0 NaN
a2 a3 NaN NaN 2.0 NaN
a4 NaN NaN 1.0 NaN
a5 NaN NaN NaN 1.0
a3 a4 1.0 NaN NaN NaN
================================================================================
Example 1
================================================================================
Result
class RR RS SR SS MIS
ax ay
a1 a2 NaN 1.0 1.0 2.0 0.0
a3 NaN NaN 2.0 NaN 0.0
a4 NaN NaN 1.0 NaN 0.0
a2 a3 NaN NaN 2.0 NaN 0.0
a4 NaN NaN 1.0 NaN 0.0
a5 NaN NaN NaN 1.0 0.0
a3 a4 1.0 NaN NaN NaN 0.0
'\n# Load data\n#data = load_susceptibility_mimic()\ndata = load_susceptibility_nhs()\n\n# Create combo\nc = create_combinations_v1(data,\n col_specimen=\'specimen_code\',\n col_lab_id=\'laboratory_number\',\n col_microorganism=\'microorganism_code\',\n col_antimicrobial=\'antimicrobial_code\',\n col_result=\'sensitivity\')\n\n# Create folder if it does not exist.\ntoday = datetime.now().strftime("%Y%m%d-%H%M%S")\npath = Path(\'./outputs/cri/\') / today\nPath(path).mkdir(parents=True, exist_ok=True)\n\n# Save combinations file.\nc.to_csv(path / \'combinations.csv\')\n\n# Build contingency\nr = c.groupby([\'specimen\', \'o\', \'ax\', \'ay\', \'class\']).size().unstack()\n\n# Compute CRI\nr[\'MIS\'] = r.fillna(0) .apply(CRI, args=(mutual_info_matrix_v3,), axis=1)\n\n# Show\nprint("\n" + "="*80 + "\nExample 2\n" + "="*80)\nprint("\nResult")\nprint(r)\n\n# Save collateral sensitivity index file.\nr.to_csv(path / \'contingency.csv\')\n'
Total running time of the script: ( 0 minutes 0.194 seconds)