Note
Click here to download the full example code
12b. Matplotlib to Plotly (shap)
This example converts a matplotlib figure to Plotly.
Warning
A known bug in Plotly (see GitHub issue) causes the mpl_to_plotly() function to fail because it references an outdated function from Matplotlib. The current fix is to downgrade to an older version of Matplotlib or to recreate your figure in Plotly manually.
[ISSUE]: https://github.com/plotly/plotly.py/issues/3624#issuecomment-1161805210 In the latest commit of plotly packages/python/plotly/plotly/matplotlylib/mpltools.py line 368, it still calls is_frame_like() function.
Note
In the latest commit of plotly packages/python/plotly/plotly/matplotlylib/mpltools.py line 368, it still calls is_frame_like() function. There is already an issue tracking this. You may need choose to downgrade Matplotlib if you still want to use mpl_to_plotly() function.
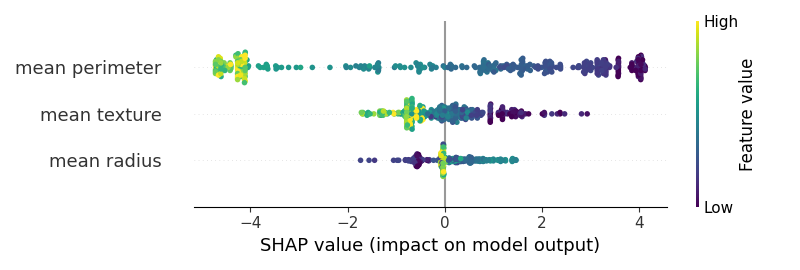
Out:
Kernel type: <class 'shap.explainers._tree.TreeExplainer'>
Shap values:
.values =
array([[-0.07339976, 1.23534454, -4.69448856],
[-0.28925516, -0.13902985, -4.83976683],
[-0.23044579, -0.75952114, -4.40960412],
...,
[ 0.41172362, 0.30341838, 1.756622 ],
[-0.28925516, -0.13902985, -4.83976683],
[-0.23044579, -0.75952114, -4.40960412]])
.base_values =
array([0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342,
0.80274342, 0.80274342, 0.80274342, 0.80274342, 0.80274342])
.data =
array([[ 17.99, 10.38, 122.8 ],
[ 20.57, 17.77, 132.9 ],
[ 19.69, 21.25, 130. ],
...,
[ 12.47, 17.31, 80.45],
[ 18.49, 17.52, 121.3 ],
[ 20.59, 21.24, 137.8 ]])
(500, 3)
(500,)
(500, 3)
'\n# Convert to plotly\nimport plotly.tools as tls\nimport plotly.graph_objs as go\n\n# Get current figure and convert\nfig = tls.mpl_to_plotly(plt.gcf())\n\n# Format\n# Update layout\nfig.update_layout(\n #xaxis_title=\'False Positive Rate\',\n #yaxis_title=\'True Positive Rate\',\n #yaxis=dict(scaleanchor="x", scaleratio=1),\n #xaxis=dict(constrain=\'domain\'),\n width=700, height=350,\n #legend=dict(\n # x=1.0, y=0.0, # x=1, y=1.02\n # orientation="v",\n # font=dict(\n # size=12,\n # color=\'black\'),\n # yanchor="bottom",\n # xanchor="right",\n #),\n font=dict(\n size=15,\n #family="Times New Roman",\n #color="black",\n ),\n title=dict(\n font=dict(\n # family="Times New Roman",\n # color="black"\n )\n ),\n yaxis=dict(\n tickmode=\'array\',\n tickvals=[0, 1, 2],\n ticktext=features[:-1],\n tickfont=dict(size=15)\n ),\n xaxis=dict(\n tickfont=dict(size=15)),\n #margin={\n # \'l\': 0,\n # \'r\': 0,\n # \'b\': 0,\n # \'t\': 0,\n # \'pad\': 4\n #},\n paper_bgcolor=\'rgba(0,0,0,0)\', # transparent\n plot_bgcolor=\'rgba(0,0,0,0)\', # transparent\n template=\'simple_white\'\n)\n\n# Update scatter\nfig.update_traces(marker={\'size\': 10})\n\n# Add vertical lin\nfig.add_vline(x=0.0, line_width=2,\n line_dash="dash", line_color="black") # green\n\n# .. note:: Would it be possible to get the values of\n# cmin, cmax and the tick vals from the shap\n# values? Ideally we do not want to hardcode\n# them.\n\n# Add colorbar\ncolorbar_trace = go.Scatter(\n x=[None], y=[None], mode=\'markers\',\n marker=dict(\n colorscale=\'viridis\',\n showscale=True,\n cmin=-5,\n cmax=5,\n colorbar=dict(thickness=20,\n tickvals=[-5, 5],\n ticktext=[\'Low\', \'High\'],\n outlinewidth=0)),\n hoverinfo=\'none\'\n)\nfig[\'layout\'][\'showlegend\'] = False\nfig.add_trace(colorbar_trace)\n\n# Show\n#fig.show()\nfig\n'
22 # Generic
23 import numpy as np
24 import pandas as pd
25 import matplotlib.pyplot as plt
26
27 # Sklearn
28 from sklearn.model_selection import train_test_split
29 from sklearn.datasets import load_iris
30 from sklearn.datasets import load_breast_cancer
31 from sklearn.naive_bayes import GaussianNB
32 from sklearn.linear_model import LogisticRegression
33 from sklearn.tree import DecisionTreeClassifier
34 from sklearn.ensemble import RandomForestClassifier
35
36 # Xgboost
37 from xgboost import XGBClassifier
38
39 try:
40 __file__
41 TERMINAL = True
42 except:
43 TERMINAL = False
44
45
46 # ----------------------------------------
47 # Load data
48 # ----------------------------------------
49 # Seed
50 seed = 0
51
52 # Load dataset
53 bunch = load_iris()
54 bunch = load_breast_cancer()
55
56 # Features
57 features = list(bunch['feature_names'])
58
59 # Create DataFrame
60 data = pd.DataFrame(data=np.c_[bunch['data'],
61 bunch['target']], columns=features + ['target'])
62
63 # Create X, y
64 X = data[bunch['feature_names']]
65 y = data['target']
66
67 # Filter
68 X = X.iloc[:500, :3]
69 y = y.iloc[:500]
70
71 # Split dataset
72 X_train, X_test, y_train, y_test = \
73 train_test_split(X, y, random_state=seed)
74
75
76 # ----------------------------------------
77 # Classifiers
78 # ----------------------------------------
79 # Define some classifiers
80 gnb = GaussianNB()
81 llr = LogisticRegression()
82 dtc = DecisionTreeClassifier(random_state=seed)
83 rfc = RandomForestClassifier(random_state=seed)
84 xgb = XGBClassifier(
85 min_child_weight=0.005,
86 eta= 0.05, gamma= 0.2,
87 max_depth= 4,
88 n_estimators= 100)
89
90 # Select one
91 clf = xgb
92
93 # Fit
94 clf.fit(X_train, y_train)
95
96 # ----------------------------------------
97 # Compute shap values
98 # ----------------------------------------
99 # Import
100 import shap
101
102 # Get generic explainer
103 explainer = shap.Explainer(clf, X_train)
104
105 # Show kernel type
106 print("\nKernel type: %s" % type(explainer))
107
108 # Get shap values
109 shap_values = explainer(X)
110
111 # Show shap values
112 print("Shap values:")
113 print(shap_values)
114 print(shap_values.values.shape)
115 print(shap_values.base_values.shape)
116 print(shap_values.data.shape)
117
118 # Get matplotlib figure
119 plot_summary = shap.summary_plot( \
120 explainer.shap_values(X_train),
121 X_train, cmap='viridis',
122 show=False)
123
124 # Show
125 #plt.show()
126
127 """
128 # Convert to plotly
129 import plotly.tools as tls
130 import plotly.graph_objs as go
131
132 # Get current figure and convert
133 fig = tls.mpl_to_plotly(plt.gcf())
134
135 # Format
136 # Update layout
137 fig.update_layout(
138 #xaxis_title='False Positive Rate',
139 #yaxis_title='True Positive Rate',
140 #yaxis=dict(scaleanchor="x", scaleratio=1),
141 #xaxis=dict(constrain='domain'),
142 width=700, height=350,
143 #legend=dict(
144 # x=1.0, y=0.0, # x=1, y=1.02
145 # orientation="v",
146 # font=dict(
147 # size=12,
148 # color='black'),
149 # yanchor="bottom",
150 # xanchor="right",
151 #),
152 font=dict(
153 size=15,
154 #family="Times New Roman",
155 #color="black",
156 ),
157 title=dict(
158 font=dict(
159 # family="Times New Roman",
160 # color="black"
161 )
162 ),
163 yaxis=dict(
164 tickmode='array',
165 tickvals=[0, 1, 2],
166 ticktext=features[:-1],
167 tickfont=dict(size=15)
168 ),
169 xaxis=dict(
170 tickfont=dict(size=15)),
171 #margin={
172 # 'l': 0,
173 # 'r': 0,
174 # 'b': 0,
175 # 't': 0,
176 # 'pad': 4
177 #},
178 paper_bgcolor='rgba(0,0,0,0)', # transparent
179 plot_bgcolor='rgba(0,0,0,0)', # transparent
180 template='simple_white'
181 )
182
183 # Update scatter
184 fig.update_traces(marker={'size': 10})
185
186 # Add vertical lin
187 fig.add_vline(x=0.0, line_width=2,
188 line_dash="dash", line_color="black") # green
189
190 # .. note:: Would it be possible to get the values of
191 # cmin, cmax and the tick vals from the shap
192 # values? Ideally we do not want to hardcode
193 # them.
194
195 # Add colorbar
196 colorbar_trace = go.Scatter(
197 x=[None], y=[None], mode='markers',
198 marker=dict(
199 colorscale='viridis',
200 showscale=True,
201 cmin=-5,
202 cmax=5,
203 colorbar=dict(thickness=20,
204 tickvals=[-5, 5],
205 ticktext=['Low', 'High'],
206 outlinewidth=0)),
207 hoverinfo='none'
208 )
209 fig['layout']['showlegend'] = False
210 fig.add_trace(colorbar_trace)
211
212 # Show
213 #fig.show()
214 fig
215 """
Total running time of the script: ( 0 minutes 1.883 seconds)